Wrangling Messy Data Files
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Alteryx Technical Whitepaper
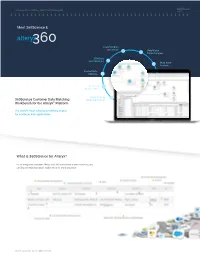
Showcase Solving the Problem - Not the Process 360Science for Alteryx Technical Whitepaper Meet 360Science & alteryx Customer Data Integration Unify Match Merge Suppress CRM Data Lead Matching Multi-buyer Analysis Contact Data Blending Customer Segmentation Cross-Channel Marketing Analysis The world’s most advanced matching engine for customer data applications. What is 360Science for Alteryx? ©2018 360Science Inc. All rights reserved Now Available! 360Science for Alteryx. It will forever change how you do customer data analytics Showcase Solving the Problem - Not the Process alteryx Technical Whitepaper Alteryx Showcase Solving the Problem - Not the Process Matching Engine Core Capabilities - Data Matching INSPECT RESULTS It all starts with the matching engine! Alteryx Traditional Customer Data Matching Process Our team of engineers, data scientists and developers DIAGNOSE have created AND perfected the industry’s most accurate BLOCK/GROUP false-positives and effective contact data matching engine. CLUSTER false-negatives MATCH KEYS missed matches > ANALYZE CREATE RETUNE/REBUILD GENERATE DEPLOY DATA SOURCES TARGET DATASET MATCH MODELS MATCH KEYS MATCHKEYS Data Source Data Wrangling > Data Modeling DEFINE MATCH REQUIREMENTS alteryx parse clean extract correct join data normalize blend data standardize Handling Set Weights & from Libraries Build and Train Experiment with Match Strategies Determine Outlier Data Dictionaries Score Thresholds Choose Algorithm MATCHCODE: First_Name(1) + Metaphone_Last_Name(5) + Street_Number(4) + ZIP(5) MATCHKEY: 360Science INTELLIGENTLY Scores Matches! Conventional solutions do not! Match/Unify Accuracy Matters! 226% Data Matching Normalization Independent testing by one of our partners reported, 360Science Matching Engine delivered up to a 226% more accurate match rate on customer CRM data than competing solutions. Geolocation Normalization Merge Transliteration Suppress Address Correction Dedupe Geolocation ©2018 360Science Inc. -

Data Cleaning Tools
A Survey of Data Cleaning Tools Group 1 Lukas Bodner, Daniel Geiger, and Lorenz Leitner 706.057 Information Visualisation SS 2020 Graz University of Technology 20 May 2020 Abstract Clean data is important to achieve correct results and prevent wrong conclusions. However, many data sources contain “unclean” data, such as improperly formatted cells, inconsistent spellings, wrong character encodings, unnecessary entries, and so on. Ideally, all these and more should be cleaned or removed before starting any analysis and work with the data. Doing this manually is tedious and often impossible, depending on the size of the data set. Therefore, data cleaning tools exist, which aim to simplify and automate these tasks, to arrive at robust and concise clean data quickly. This survey looks at different solutions for data cleaning, in specific multiple data cleaning tools. They are described, tested and evaluated, using the same unclean data sets to provide consistency. A feature matrix is created which shows an overview of all data cleaning tools and their characteristics, which can be used for a quick comparison. Some of the tools are described and tested in more detail. A final general recommendation is also provided. © Copyright 2020 by the author(s), except as otherwise noted. This work is placed under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence. Contents Contents ii List of Figures iii List of Tables v List of Listings vii 1 Introduction 1 2 Data Sets 3 2.1 Data Set 1: Parking Spots in Graz (Task: Merging) . .3 2.2 Data Set 2: Candy Ratings (Task: Standardization) . -

Integrating High-Performance Machine Learning: H2O and KNIME
Integrating high-performance machine learning: H2O and KNIME Mark Landry (H2O), Christian Dietz (KNIME) © 2017 KNIME AG. All Rights Reserved. Speed Accuracy H2O: in-memory machine learning platform designed for speed on distributed systems 2 High Level Architecture HDFS H2O Compute Engine Exploratory & Supervised & S3 Load Data Descriptive Unsupervised Predict Analysis Modeling Distributed In-Memory Feature Model NFS Data & Model Engineering & Evaluation & Loss-less Storage Compression Selection Selection Local Data Prep Export: Model Export: Plain Old Java Plain Old Java Object Object SQL Production Scoring Environment Your Imagination 3 Distributed Algorithms Advantageous Foundation • Foundation for In-Memory Distributed Algorithm Calculation - Distributed Data Frames and columnar compression • All algorithms are distributed in H2O: GBM, GLM, DRF, Parallel Parse into Distributed Rows Deep Learning and more. Fine-grained map-reduce iterations. • Only enterprise-grade, open-source distributed algorithms in the market User Benefits • “Out-of-box” functionalities for all algorithms (NO MORE SCRIPTING) and uniform interface across all languages: R, Python, Java • Designed for all sizes of data sets, especially large data Foundation for Distributed Algorithms Distributed for Foundation • Highly optimized Java code for model exports Fine Grain Map Reduce Illustration: Scalable • In-house expertise for all algorithms Distributed Histogram Calculation for GBM 4 Scientific Advisory Council 5 Machine Learning Benchmarks (https://github.com/szilard/benchm-ml) Time (s) AUC X = million of samples X = million of samples Gradient Boosting Machine Benchmark (also available for GLM and Random Forest) 6 H2O Algorithms Supervised Learning Unsupervised Learning • Generalized Linear Models: Binomial, • K-means: Partitions observations into k Statistical Gaussian, Gamma, Poisson and Tweedie Clustering clusters/groups of the same spatial size. -

Introducing Data Science to Undergraduates Through Big Data: Answering Questions by Wrangling and Profiling a Yelp Dataset
Proceedings of the 50th Hawaii International Conference on System Sciences | 2017 Introducing Data Science to Undergraduates through Big Data: Answering Questions by Wrangling and Profiling a Yelp Dataset Scott Jensen Lucas College and Graduate School of Business, San José State University [email protected] Abstract formulate data questions, and domain business There is an insatiable demand in industry for data knowledge. In a recent study of analytics and data scientists, and graduate programs and certificates science programs at the undergraduate level, are gearing up to meet this demand. However, there Aasheim et al. [1] found that data mining and is agreement in the industry that 80% of a data analytics/modeling were covered in 100% of such scientist’s work consists of the transformation and programs. Of the four major areas reviewed, the area profiling aspects of wrangling Big Data; work that least frequently covered was Big Data. However, may not require an advanced degree. In this paper Big Data is the fuel that powers data science. we present hands-on exercises to introduce Big Data Undergraduate students could be introduced to data to undergraduate MIS students using the CoNVO science by learning to wrangle a dataset and answer Framework and Big Data tools to scope a data questions using Big Data tools. Since a key aspect of problem and then wrangle the data to answer data science is learning to decide what questions to questions using a real world dataset. This can ask of a dataset, students also need to learn to provide undergraduates with a single course formulate their own questions. -

Cheat Sheet: Data Wrangling with KNIME Analytics Platform
Cheat Sheet: Data Wrangling with KNIME Analytics Platform ACCESS DATA COMBINE DATA FILTER DATA WRITE DATA DATE&TIME Row Filter CSV Reader Reads a CSV file from either your local Amazon S3 Concatenates the Connector Connects to Amazon S3 and points to a Filters rows in or out of the input table Writes the input table(s) to sheet(s) Parses the strings in the selected file system or another connected file rows of all input working directory (with a UNIX-like syntax, e.g., Concatenate according to a filtering rule. The filtering rule Excel Writer in an Excel file (XLS or XLSX). Click String to Date&Time columns according to a date/time system. Click the three dots in the lower tables by writing /mybucket/myfolder/myfile). Allows can match a value in a selected column or the three dots in the lower left format and converts them into left corner to add a dynamic connection them below each downstream reader nodes to access data from numbers in a numerical range. corner to add a dynamic sheet input Date&Time cells. Four Date&Time input port to connect to an external file other. This is Amazon S3 as a file system. port to write multiple data tables forms are supported: only date, only system, like Amazon S3, Azure Blob especially useful for Rule-based Row Filter into multiple sheets. time, date&time, and date&time plus Storage, etc. tables with shared Filters rows in or out according to a set of rules, defined in its configuration window. time zone. -

Trifacta Architecture: an Intelligent Data Wrangling Platform
575 Market St, 11th Floor trifacta.com San Francisco, CA 94105 844.332.2821 WHITEPAPER Trifacta Architecture: An Intelligent & Interoperable Data Wrangling Platform Trifacta was born out of the belief that the people who know the data best should be able to wrangle it themselves. Instead of a rigid and highly-technical data transformation process, we sought out to build an intuitive user experience and workflow for every user within an organization. This required years of research at UC Berkeley and Stanford across machine learning, human-computer interaction and parallel processing, a deep consideration for the analytics ecosystem in its entirety, and an enduring empathy for end-user analysts, which eventually produced the world’s leading data platform. Today, Trifacta’s architecture is validated by leading industry analysts and in production at industry-leading organizations around the world. Trifacta sits between data storage and processing environments and the visualization, statistical or machine learning tools used downstream. As a best-in-breed technology, Trifacta has been architected to be open and adaptable so as the technologies upstream and downstream change, the investments and logic created in Trifacta are able to utilize those innovations. What follows are foundational aspects of Trifacta. ©2017 Trifacta . All rights reserved. Trifacta Architecture 2 APPLICATIONS Publishing & Access Framework Core Data Wrangling User Experience Wrangle Language Intelligence & Context Governance Any Scale Data Processing Collaboration Operationalization Metadata Management Data Security & Administration Connectivity Framework DATA SOURCES Ecosystem & Extensibility Connectivity Framework Trifacta maintains a robust connectivity and API framework, enabling users to access live data without requiring them to pre-load or create a copy of the data separate from the source data system. -

QBS 181: Data Wrangling
QBS 181: Data Wrangling Fall 2020 Tuesday and Thursday, TBD Course Instructor: Ramesh Yapalparvi Data Wrangling: This course introduces techniques which enables students to pull data from various sources and to work with different forms of data. Data wrangling requires a strong knowledge of the data structures that hold datasets. This course will cover the different types of data structures available in SQL and R, how they differ by dimensionality and how to create, add to, and subset the various data structures. You will also learn how to deal with missing values in data structures. Furthermore, since analysis is often a collaborative effort data scientist also need to know how to share their data. This course will cover the basics of importing tabular and spreadsheet data, scraping data stored online, and exporting data for sharing purposes. Writing a simple, replicable, and readable code is important to becoming an effective and efficient data scientist. Throughout this course, you will be introduced to the art of writing functions and using loop control statements to reduce redundancy in code. You will also learn how to simplify your code using various operators to make your code more readable. Consequently, this will help students to perform data wrangling tasks more effectively, efficiently, and with more clarity. Data wrangling is all about getting your data into the right form in order to feed it into the visualization and modeling stages. This typically requires a large amount of reshaping and transforming of your data. Throughout this course, you will learn the fundamental functions for “tidying” your data and for manipulating, sorting, summarizing, and joining your data. -

Six Core Data Wrangling Activities
Six Core Data Wrangling Activities An introductory guide to data wrangling with Trifacta Today’s Data Driven Culture Are you inundated with data? Today, most organizations are collecting as much data in one week as they used to collect in one year. Even though they have more data, they’re not using the bulk of it —most companies analyze less than 20% of their data. Why? Preparing diverse datasets for analysis is time-consuming and messy, often left to data engineers or data scientists. 2 Introducing, Trifacta Wrangler! With Trifacta Wrangler, analysts, researchers and data visualization specialists no longer need to be dependent on IT for data preparation. Trifacta empowers you to wrangle data yourself — allowing you to spend less time waiting and more time on your analysis. Start wrangling your data by following the 6 steps of the process: Discovering Structuring Cleaning Enriching Validating Publishing 3 Discovering Structuring Cleaning Enriching Validating Publishing All Data Wrangling with Trifacta A step-by-step walk-through using Bay Area Bike Share data Each of the six data wrangling steps will be demonstrated using Trifacta to wrangle Bay Area Bike Share and weather data, dating from August 29, 2014 to September 2, 2014 and the entire year of 2014 respectively. The Bay Area Bike Share is a program that includes 700 bicycles across 70 stations in the Bay Area for the purpose of providing residents and visitors transportation for going to work, to the CalTrain station, and generally getting around. An analyst who works for Bay Area Bike Share might want to examine the data in order to see the most popular bike routes and stations to better plan bike inventory and rotations. -

Introduction to R: Data Wrangling
Introduction to R: Data Wrangling Lisa Federer, Research Data Informationist March 28, 2016 This course is designed to give you a simple and easy introduction to R, a programming language that can be used for data wrangling and processing, statistical analysis, visualization, and more. This handout will walk you through every step of today's class. Throughout the handout, you'll see the example code displayed like this: > print(2 + 2) [1] 4 The part that is in italics and preceded by the > symbol is the code that you will type. The part below it is the output you should expect to see. Sometimes code doesn't have any output to print; in that case, you'll just see the code and nothing else. Also, sometimes the code is too long to fit on a single line of text in this handout. When that is the case, you will see the code split into separate lines, each starting with a +, like this: > long_line_of_code <- c("Some really long code", "oh my gosh, how long is it going to be?", + "is it going to go on forever?", "I don't know, AGGGHHHHH", + "please, make it stop!") When this is the case, do not insert any line breaks, extra spaces, or the plus sign - your code should be typed as one single line of code. Note that the default for your display in R Studio is not to wrap lines of text in your code, but you can turn this on by going to Tools > Global Options > Code Editing, and check the box next to "Soft-wrap R source files." 1 Getting Started In this class, we'll be using RStudio, which is what's known as an integrated development environment (IDE). -
Titelfolie Mit Langer Überschrift, Zwei- Oder Mehrzeilig, TT Norms Pro, 28 Pt
A visual programming approach for intuitive handling of bioprocess data Robert Söldner,*1, Jonas Austerjost1, Simon Stumm1, David J. Pollard1 1 Sartorius AG, Corporate Research, August-Spindler-Straße 11, 37097 * Corresponding author: [email protected] Introduction This simplification process allows for the easy combination of workflow steps into “macro” elements, which enhances the clarity of the During the last decade the expansion of high throughput biology and application. Several of these overarching macro elements were automated bioprocessing workflows has progressed by the advances in implemented. These included the transfer of specific measured robotics, automation and sensor technology. As a result, the size and parameters (DO, pH, current volume) of a specific fermentation vessel number of datasets dramatically grew as did the complexity. An into designated tables, as well as string to number conversion upstream bioprocess engineer or scientist may have to manipulate & combined with a forward fill of the imported raw data. wrangle datasets from several different sources, such as online Furthermore, data visualization features were introduced as part of the bioreactor profiles, and offline sample analysis, to process one typical data processing “assembly line”. Again, the basic components of experiment. This data wrangling burden to the end users often KNIME were modified to fit the demanded visualization application counteracts any efficiency advantage from using high throughput tools. (see Figure 2). This generated a requirement for data aware scientists to intervene over the wrangling and processing of complex datasets. This inefficient approach desperately needs a streamlined workflow to eliminate the need of experts for repetitive data processing tasks. An innovative approach to simplify and democratize basic data processing tasks is the introduction of visual programming. -

SAS® Visual Data Mining and Machine Learning Everything Needed to Solve Your Most Complex Problems Within a Single, Integrated In-Memory Environment
FACT SHEET SAS® Visual Data Mining and Machine Learning Everything needed to solve your most complex problems within a single, integrated in-memory environment What does SAS® Visual Data Mining and Machine Learning do? SAS Visual Data Mining and Machine Learning combines data wrangling, data explora- tion, visualization, feature engineering, and modern statistical, data mining and machine- learning techniques all in a single, integrated in-memory processing environment. This provides faster, more accurate answers to complex business problems, increased deploy- ment flexibility, and reliable, secure analytics management and governance for agile IT. Why is SAS® Visual Data Mining and Machine Learning important? It enables data scientists and others to solve previously unfeasible business problems by removing barriers created by data sizes, data diversity, limited analytical depth and computational bottlenecks. Dramatic performance gains and innovative algorithms mean greater productivity and faster, more creative answers to your most complex problems. For whom is SAS® Visual Data Mining and Machine Learning designed? It is designed for those who want to use powerful and customizable in-memory algorithms in interactive and programming interfaces to analyze large, complicated data and uncover new insights faster. This includes business analysts, data scientists, experienced statisti- cians, data miners, engineers, researchers and scientists. Data volumes continue to Benefits find the best performing model and grow. Highly skilled data provide answers with high levels of • Solve complex analytical problems scientists and analytical confidence. faster. This solution runs on SAS® Viya™, professionals are in short • Empower users with language options. the latest addition to the SAS Platform, supply. Organizations Python, R, Java and Lua programmers delivering predictive modeling and struggle to find timely answers to increas- can experience the power of this machine-learning capabilities at break- ingly complex problems. -

Inventory Management Automotive Industry KNIME Success Story
This Success Story was written by our partner: KNIME Success Story Avoiding losses from obsolete inventory Automotive Inventory Management Predictive Analytics Replacing an inefficient, cumbersome and manual two-week process Automotive Holdings Group (AHG) is a diversified automotive retailing and logistics group headquartered in Perth, Western Australia. As Australia’s largest motor dealership group, AHG owns over 180 car and truck franchises and offers a wide range of services including finance, after care, insurance and equipment. The National Trucks Division regularly undertakes analysis to identify opportunities to transfer spare parts between dealerships in order to minimise the write-downs that occur when parts age without being utilised. Historically the analysis was completed on a monthly basis via an inefficient, cumbersome and manual two-week process that originated with CSV extracts. AHG needed a scalable, flexible, transparent, and easy-to-update solution was needed, which also significantly accelerated the time to insight. An end-to-end, automated solution Results Forest Grove developed an end-to-end automated data wrangling and reporting engine to This solution has vastly expanded AHG’s deep analysis expand AHG’s capabilities in deep analysis through the process of data extraction, capabilties and represents a major milestone in its transformation, reporting and visualization. The solution is scalable, flexible and transparent, journey from descriptive to predictive analytics. By and can be easily updated to suit changes to the data environment and user requirements. It automating the data interpretation process with comprises five components: KNIME, managers are now able to identify potential transfer opportunities in minutes rather than weeks 1.