Spiral Structure of Escherichia Coli HU Provides Foundation for DNA Supercoiling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Study of Spiral Transition Curves As Related to the Visual Quality of Highway Alignment
A STUDY OF SPIRAL TRANSITION CURVES AS RELA'^^ED TO THE VISUAL QUALITY OF HIGHWAY ALIGNMENT JERRY SHELDON MURPHY B, S., Kansas State University, 1968 A MJvSTER'S THESIS submitted in partial fulfillment of the requirements for the degree MASTER OF SCIENCE Department of Civil Engineering KANSAS STATE UNIVERSITY Manhattan, Kansas 1969 Approved by P^ajQT Professor TV- / / ^ / TABLE OF CONTENTS <2, 2^ INTRODUCTION 1 LITERATURE SEARCH 3 PURPOSE 5 SCOPE 6 • METHOD OF SOLUTION 7 RESULTS 18 RECOMMENDATIONS FOR FURTHER RESEARCH 27 CONCLUSION 33 REFERENCES 34 APPENDIX 36 LIST OF TABLES TABLE 1, Geonetry of Locations Studied 17 TABLE 2, Rates of Change of Slope Versus Curve Ratings 31 LIST OF FIGURES FIGURE 1. Definition of Sight Distance and Display Angle 8 FIGURE 2. Perspective Coordinate Transformation 9 FIGURE 3. Spiral Curve Calculation Equations 12 FIGURE 4. Flow Chart 14 FIGURE 5, Photograph and Perspective of Selected Location 15 FIGURE 6. Effect of Spiral Curves at Small Display Angles 19 A, No Spiral (Circular Curve) B, Completely Spiralized FIGURE 7. Effects of Spiral Curves (DA = .015 Radians, SD = 1000 Feet, D = l** and A = 10*) 20 Plate 1 A. No Spiral (Circular Curve) B, Spiral Length = 250 Feet FIGURE 8. Effects of Spiral Curves (DA = ,015 Radians, SD = 1000 Feet, D = 1° and A = 10°) 21 Plate 2 A. Spiral Length = 500 Feet B. Spiral Length = 1000 Feet (Conpletely Spiralized) FIGURE 9. Effects of Display Angle (D = 2°, A = 10°, Ig = 500 feet, = SD 500 feet) 23 Plate 1 A. Display Angle = .007 Radian B. Display Angle = .027 Radiaji FIGURE 10. -

Construction Surveying Curves
Construction Surveying Curves Three(3) Continuing Education Hours Course #LS1003 Approved Continuing Education for Licensed Professional Engineers EZ-pdh.com Ezekiel Enterprises, LLC 301 Mission Dr. Unit 571 New Smyrna Beach, FL 32170 800-433-1487 [email protected] Construction Surveying Curves Ezekiel Enterprises, LLC Course Description: The Construction Surveying Curves course satisfies three (3) hours of professional development. The course is designed as a distance learning course focused on the process required for a surveyor to establish curves. Objectives: The primary objective of this course is enable the student to understand practical methods to locate points along curves using variety of methods. Grading: Students must achieve a minimum score of 70% on the online quiz to pass this course. The quiz may be taken as many times as necessary to successful pass and complete the course. Ezekiel Enterprises, LLC Section I. Simple Horizontal Curves CURVE POINTS Simple The simple curve is an arc of a circle. It is the most By studying this course the surveyor learns to locate commonly used. The radius of the circle determines points using angles and distances. In construction the “sharpness” or “flatness” of the curve. The larger surveying, the surveyor must often establish the line of the radius, the “flatter” the curve. a curve for road layout or some other construction. The surveyor can establish curves of short radius, Compound usually less than one tape length, by holding one end Surveyors often have to use a compound curve because of the tape at the center of the circle and swinging the of the terrain. -

The Ordered Distribution of Natural Numbers on the Square Root Spiral
The Ordered Distribution of Natural Numbers on the Square Root Spiral - Harry K. Hahn - Ludwig-Erhard-Str. 10 D-76275 Et Germanytlingen, Germany ------------------------------ mathematical analysis by - Kay Schoenberger - Humboldt-University Berlin ----------------------------- 20. June 2007 Abstract : Natural numbers divisible by the same prime factor lie on defined spiral graphs which are running through the “Square Root Spiral“ ( also named as “Spiral of Theodorus” or “Wurzel Spirale“ or “Einstein Spiral” ). Prime Numbers also clearly accumulate on such spiral graphs. And the square numbers 4, 9, 16, 25, 36 … form a highly three-symmetrical system of three spiral graphs, which divide the square-root-spiral into three equal areas. A mathematical analysis shows that these spiral graphs are defined by quadratic polynomials. The Square Root Spiral is a geometrical structure which is based on the three basic constants: 1, sqrt2 and π (pi) , and the continuous application of the Pythagorean Theorem of the right angled triangle. Fibonacci number sequences also play a part in the structure of the Square Root Spiral. Fibonacci Numbers divide the Square Root Spiral into areas and angle sectors with constant proportions. These proportions are linked to the “golden mean” ( golden section ), which behaves as a self-avoiding-walk- constant in the lattice-like structure of the square root spiral. Contents of the general section Page 1 Introduction to the Square Root Spiral 2 2 Mathematical description of the Square Root Spiral 4 3 The distribution -

Archimedean Spirals ∗
Archimedean Spirals ∗ An Archimedean Spiral is a curve defined by a polar equation of the form r = θa, with special names being given for certain values of a. For example if a = 1, so r = θ, then it is called Archimedes’ Spiral. Archimede’s Spiral For a = −1, so r = 1/θ, we get the reciprocal (or hyperbolic) spiral. Reciprocal Spiral ∗This file is from the 3D-XploreMath project. You can find it on the web by searching the name. 1 √ The case a = 1/2, so r = θ, is called the Fermat (or hyperbolic) spiral. Fermat’s Spiral √ While a = −1/2, or r = 1/ θ, it is called the Lituus. Lituus In 3D-XplorMath, you can change the parameter a by going to the menu Settings → Set Parameters, and change the value of aa. You can see an animation of Archimedean spirals where the exponent a varies gradually, from the menu Animate → Morph. 2 The reason that the parabolic spiral and the hyperbolic spiral are so named is that their equations in polar coordinates, rθ = 1 and r2 = θ, respectively resembles the equations for a hyperbola (xy = 1) and parabola (x2 = y) in rectangular coordinates. The hyperbolic spiral is also called reciprocal spiral because it is the inverse curve of Archimedes’ spiral, with inversion center at the origin. The inversion curve of any Archimedean spirals with respect to a circle as center is another Archimedean spiral, scaled by the square of the radius of the circle. This is easily seen as follows. If a point P in the plane has polar coordinates (r, θ), then under inversion in the circle of radius b centered at the origin, it gets mapped to the point P 0 with polar coordinates (b2/r, θ), so that points having polar coordinates (ta, θ) are mapped to points having polar coordinates (b2t−a, θ). -

The Logarithmic Spiral * the Parametric Equations for The
The Logarithmic Spiral * The parametric equations for the Logarithmic Spiral are: x(t) =aa exp(bb t) cos(t) · · · y(t) =aa exp(bb t) sin(t). · · · This spiral is connected with the complex exponential as follows: x(t) + i y(t) = aa exp((bb + i)t). The animation that is automatically displayed when you select Logarithmic Spiral from the Plane Curves menu shows the osculating circles of the spiral. Their midpoints draw another curve, the evolute of this spiral. These os- culating circles illustrate an interesting theorem, namely if the curvature is a monotone function along a segment of a plane curve, then the osculating circles are nested - because the distance of the midpoints of two osculating circles is (by definition) the length of a secant of the evolute while the difference of their radii is the arc length of the evolute between the two midpoints. (See page 31 of J.J. Stoker’s “Differential Geometry”, Wiley-Interscience, 1969). For the logarithmic spiral this implies that through every point of the plane minus the origin passes exactly one os- culating circle. Etienne´ Ghys pointed out that this leads * This file is from the 3D-XplorMath project. Please see: http://3D-XplorMath.org/ 1 to a surprise: The unit tangent vectors of the osculating circles define a vector field X on R2 0 – but this vec- tor field has more integral curves, i.e.\ {solution} curves of the ODE c0(t) = X(c(t)), than just the osculating circles, namely also the logarithmic spiral. How is this compatible with the uniqueness results of ODE solutions? Read words backwards for explanation: eht dleifrotcev si ton ztihcspiL gnola eht evruc. -

Cotes's Spiral Vortex in Extratropical Cyclone Bomb South Atlantic Oceans
Cotes’s Spiral Vortex in Extratropical Cyclone bomb South Atlantic Oceans Ricardo Gobato1,*, Alireza Heidari2, Abhijit Mitra3 and Marcia Regina Risso Gobato1 1Green Land Landscaping and Gardening, Seedling Growth Laboratory, 86130-000, Parana, Brazil. 2Faculty of Chemistry, California South University, 14731 Comet St. Irvine, CA 92604, USA, United States of America. 3Department of Marine Science, University of Calcutta, 35 B.C. Road Kolkata, 700019, India. *Corresponding author: [email protected] September 20, 2020 he characteristic shape of hurricanes, cy- pressure center of 972 mbar , approximate loca- clones, typhoons is a spiral. There are sev- tion 34◦S 42◦30’W. eral types of turns, and determining the Tcharacteristic equation of which spiral the "cyclone bomb" (CB) fits into is the goal of the work. In 1 Introduction mathematics, a spiral is a curve which emanates from a point, moving farther away as it revolves In mathematics, a spiral is a curve which emanates around the point. An “explosive extratropical cy- from a point, moving farther away as it revolves around clone” is an atmospheric phenomenon that occurs the point. [1-4] The characteristic shape of hurricanes, when there is a very rapid drop in central at- cyclones, typhoons is a spiral [5-8]. There are sev- mospheric pressure. This phenomenon, with its eral types of turns, and determining the characteristic characteristic of rapidly lowering the pressure in equation of which spiral the cyclone bomb CB [9]. its interior, generates very intense winds and for The work aims to determine the mathematical equa- this reason it is called explosive cyclone, bomb cy- tion of the shape of the extratropical cyclone, in the clone. -

Chapter 2 Track
CALTRAIN DESIGN CRITERIA CHAPTER 2 - TRACK CHAPTER 2 TRACK A. GENERAL This Chapter includes criteria and standards for the planning, design, construction, and maintenance as well as materials of Caltrain trackwork. The term track or trackwork includes special trackwork and its interface with other components of the rail system. The trackwork is generally defined as from the subgrade (or roadbed or trackbed) to the top of rail, and is commonly referred to in this document as track structure. This Chapter is organized in several main sections, namely track structure and their materials including civil engineering, track geometry design, and special trackwork. Performance charts of Caltrain rolling stock are also included at the end of this Chapter. The primary considerations of track design are safety, economy, ease of maintenance, ride comfort, and constructability. Factors that affect the track system such as safety, ride comfort, design speed, noise and vibration, and other factors, such as constructability, maintainability, reliability and track component standardization which have major impacts to capital and maintenance costs, must be recognized and implemented in the early phase of planning and design. It shall be the objective and responsibility of the designer to design a functional track system that meets Caltrain’s current and future needs with a high degree of reliability, minimal maintenance requirements, and construction of which with minimal impact to normal revenue operations. Because of the complexity of the track system and its close integration with signaling system, it is essential that the design and construction of trackwork, signal, and other corridor wide improvements be integrated and analyzed as a system approach so that the interaction of these elements are identified and accommodated. -
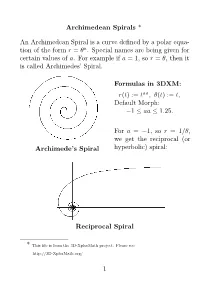
Archimedean Spirals * an Archimedean Spiral Is a Curve
Archimedean Spirals * An Archimedean Spiral is a curve defined by a polar equa- tion of the form r = θa. Special names are being given for certain values of a. For example if a = 1, so r = θ, then it is called Archimedes’ Spiral. Formulas in 3DXM: r(t) := taa, θ(t) := t, Default Morph: 1 aa 1.25. − ≤ ≤ For a = 1, so r = 1/θ, we get the− reciprocal (or Archimede’s Spiral hyperbolic) spiral: Reciprocal Spiral * This file is from the 3D-XplorMath project. Please see: http://3D-XplorMath.org/ 1 The case a = 1/2, so r = √θ, is called the Fermat (or hyperbolic) spiral. Fermat’s Spiral While a = 1/2, or r = 1/√θ, it is called the Lituus: − Lituus 2 In 3D-XplorMath, you can change the parameter a by go- ing to the menu Settings Set Parameters, and change the value of aa. You can see→an animation of Archimedean spirals where the exponent a = aa varies gradually, be- tween 1 and 1.25. See the Animate Menu, entry Morph. − The reason that the parabolic spiral and the hyperbolic spiral are so named is that their equations in polar coor- dinates, rθ = 1 and r2 = θ, respectively resembles the equations for a hyperbola (xy = 1) and parabola (x2 = y) in rectangular coordinates. The hyperbolic spiral is also called reciprocal spiral be- cause it is the inverse curve of Archimedes’ spiral, with inversion center at the origin. The inversion curve of any Archimedean spirals with re- spect to a circle as center is another Archimedean spiral, scaled by the square of the radius of the circle. -

Fermat's Spiral and the Line Between Yin and Yang
FERMAT’S SPIRAL AND THE LINE BETWEEN YIN AND YANG TARAS BANAKH ∗, OLEG VERBITSKY †, AND YAROSLAV VOROBETS ‡ Abstract. Let D denote a disk of unit area. We call a set A D perfect if it has measure 1/2 and, with respect to any axial symmetry of D⊂, the maximal symmetric subset of A has measure 1/4. We call a curve β in D an yin-yang line if β splits D into two congruent perfect sets, • β crosses each concentric circle of D twice, • β crosses each radius of D once. We• prove that Fermat’s spiral is a unique yin-yang line in the class of smooth curves algebraic in polar coordinates. 1. Introduction The yin-yang concept comes from ancient Chinese philosophy. Yin and Yang refer to the two fundamental forces ruling everything in nature and human’s life. The two categories are opposite, complementary, and intertwined. They are depicted, respectively, as the dark and the light area of the well-known yin-yang symbol (also Tai Chi or Taijitu, see Fig. 1). The borderline between these areas represents in Eastern thought the equilibrium between Yin and Yang. From the mathematical point of view, the yin-yang symbol is a bipartition of the disk by a certain curve β, where by curve we mean the image of a real segment under an injective continuous map. We aim at identifying this curve, up to deriving an explicit mathematical expression for it. Such a project should apparently begin with choosing a set of axioms for basic properties of the yin-yang symbol in terms of β. -

Mathematics Extended Essay
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by TED Ankara College IB Thesis MATHEMATICS EXTENDED ESSAY “Relationship Between Mathematics and Art” Candidate Name: Zeynep AYGAR Candidate Number: 1129-018 Session: MAY 2013 Supervisor: Mehmet Emin ÖZER TED Ankara College Foundation High School ZEYNEP AYGAR D1129018 ABSTRACT For most of the people, mathematics is a school course with symbols and rules, which they suffer understanding and find difficult throughout their educational life, and even they find it useless in their daily life. Mathematics and arts, in general, are considered and put apart from each other. Mathematics represents truth, while art represents beauty. Mathematics has theories and proofs, but art depends on personal thought. Even though mathematics is a combination of symbols and rules, it has strong effects on daily life and art. It is obvious that those people dealing with strict rules of positive sciences only but nothing else suffer from the lack of emotion. Art has no meaning for them. On the other hand, artists unaware of the rules controlling the whole universe are living in a utopic world. However, a man should be aware both of the real world and emotional world in order to become happy in his life. In order to achieve that he needs two things: mathematics and art. Mathematics is the reflection of the physical world in human mind. Art is a mirror of human spirit. While mathematics showing physical world, art reveals inner world. Then it is not wrong to say that they have strong relations since they both have great effects on human beings. -

From Spiral to Spline: Optimal Techniques in Interactive Curve Design
From Spiral to Spline: Optimal Techniques in Interactive Curve Design by Raphael Linus Levien A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Engineering–Electrical Engineering and Computer Sciences in the GRADUATE DIVISION of the UNIVERSITY OF CALIFORNIA, BERKELEY Committee in charge: Professor Carlo S´equin, Chair Professor Jonathan Shewchuk Professor Jasper Rine Fall 2009 From Spiral to Spline: Optimal Techniques in Interactive Curve Design Copyright © 2009 by Raphael Linus Levien Abstract From Spiral to Spline: Optimal Techniques in Interactive Curve Design by Raphael Linus Levien Doctor of Philosophy in Engineering–Electrical Engineering and Computer Sciences University of California, Berkeley Professor Carlo S´equin, Chair A basic technique for designing curved shapes in the plane is interpolating splines. The designer inputs a sequence of control points, and the computer fits a smooth curve that goes through these points. The literature of interpolating splines is rich, much of it based on the mathematical idealization of a thin elastic strip constrained to pass through the points. Until now there is little consensus on which, if any, of these splines is ideal. This thesis explores the properties of an ideal interpolating spline. The most important property is fairness, a property often in tension with locality, meaning that perturbations to the input points do not affect sections of the curve at a distance. The idealized elastic strip has two serious problems. A sequence of co-circular input points results in a curve deviating from a circular arc. For some other inputs, no solution (with finite extent) exists at all. -

Golden Ratio, Golden Section, Golden Mean, Golden Spiral, Phi, Geometrical Validation of Phi, Fibonacci Number, Phi in Nature, Equation of Phi
International Journal of Arts 2011; 1(1): 1-22 DOI: 10.5923/j.arts.20110101.01 Geometrical Substantiation of Phi, the Golden Ratio and the Baroque of Nature, Architecture, Design and Engineering Md. Akhtaruzzaman*, Amir A. Shafie Department of Mechatronics Engineering, Kulliyyah of Engineering, International Islamic University Malaysia, Kuala Lumpur, 53100, Malaysia Abstract Golden Proportion or Golden Ratio is usually denoted by the Greek letter Phi (φ), in lower case, which repre- sents an irrational number, 1.6180339887 approximately. Because of its unique and mystifying properties, many researchers and mathematicians have been studied about the Golden Ratio which is also known as Golden Section. Renaissance architects, artists and designers also studied on this interesting topic, documented and employed the Golden section proportions in eminent works of artifacts, sculptures, paintings and architectures. The Golden Proportion is considered as the most pleasing to human visual sensation and not limited to aesthetic beauty but also be found its existence in natural world through the body proportions of living beings, the growth patterns of many plants, insects and also in the model of enigmatic universe. The properties of Golden Section can be instituted in the pattern of mathematical series and geometrical patterns. This paper seeks to represent a panoptic view of the miraculous Golden Proportion and its relation with the nature, globe, universe, arts, design, mathematics and science. Geometrical substantiation of the equation of Phi, based on the classical geometric relations, is also explicated in this study. Golden Ratio and its chronicle, concept of Golden Mean and its relations with the geometry, various dynamic rectangles and their intimacy with Phi, Golden Ratio in the beauty of nature, Phi ratio in the design, architecture and engineering are also presented in this study in a panoptical manner.