In Silico Structural Characterization of Monooxygenase and Dioxygenase Enzymes from Danio Rerio
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Wp(C) 2184/2010
IN THE GAUHATI HIGH COURT (THE HIGH COURT OF ASSAM: NAGALAND: MEGHALAYA: MANIPUR: TRIPURA: MIZORAM AND ARUNACHAL PRADESH) WP(C) No.2184/2010 1.Shri Manabendra Sarma, Son of Sri Sarat Ch. Sarma, Resident of Durga Sarobar, Kamakhya Gate, P.O. Bharalumukh, District- Kamrup, Assam. 2.Shri Khargeswar Bora, Son of Late Nareswar Bora, Resident of Vill & P.O.-Tulashimukh, District- Nagaon, Assam. 3.Shri Mukut Deka, Son of Late Bhagawan Ch. Deka, Resident of Ananda Nagar, Bye-Lane No.1, P.O. Noonmati, District- Kamrup, Assam. 4.Shri Utpal Tamuly, Son of Sambhu Ram Tamuly, Resident of Vill-Abhaypur, P.O.- College Nagar, Guwahati-31, District- Kamrup, Assam. 5.Shri Paban Kumar Hazarika, Son of Sri Joynath Hazarika, Village-Bisoyachook, P.O. Puranigudam, District- Nagaon. 6.Shri Nipan Mahanta, S/o Sri Sarat Ch. Mahanta, Resident of Vill & P.O.-Sualkuchi, District- Kamrup, Assam. 2 7.Shri Apurba Kr. Baruah, Son of Sri BN Baruah, Resident of Village- Kachalukhowa, P.O.-Nagaon, District- Nagaon, Assam. 8.Shri Mriganka Kumar Baruah, S/o Shri Jugal Mohan Baruah, Resident of Hatigaon, Guwahati, District- Kamrup. 9.Shri Ashim Jyoti Moral, C/o Dr. Bhagaban Moral, Fatasil G.S. Colony, Guwahati, District- Kamrup. 10.Shri Jitu Moni Hazarika, C/o Shri Thuleswar Hazarika, Village & P.O.-Debnarikali, District- Nagaon. 11.Shri Pranab Kumar Sarmah, S/o Late Guna Ram Sarmah, Village & P.O.-Borangutoli, District- Nagaon. 12.Shri Iftikar Rahman, Son of Rejifur Rahman, Resident of Vill- Ghutulai Patty, Haibargaon, District- Nagaon, Assam. 13.Shri Dipjyoti Saikia, Son of Sri Munindra Nath Saikia, Resident of Town Panigaon, Nagaon, District- Nagaon, Assam. -

LOK SABHA DEBATES (English Version)
Twel ........ Vol. III, No. 20 Monday, July 13, 1. Asadha 22,1120 (Salal) LOK SABHA DEBATES (English Version) Second Session (Twelftb Lok Sabba) (Vol. //I contains Nos. 11 to 20) LOK SABHA SECRETARIAT NEW DELHI Price : Rs. 50.00 EDITORIAL BOARD Shrl S. Gopalan Secretary General Lok Sabha Dr. A.K. Pandey Additional Secretary Lok Sabha Secretariat Shrl S.K. Kauahlk Director Lok Sabha Secretariat Shrl P.C. Bhatt Chief Editor Lok Sabha Seeretarlat Shrl A.P. Chakravarti Senior Editor Lok Sabha Secretariat [Original English Proceedings Included In English Version and Original Hindi proceedings Included In Hindi Version will be treated as authoritative and not the translation thereof.] 1IcndIr, .:IJly 13, ~ 22, 1920 (SUa) ODl./line 8/23 'rIlE MINISl'ER OF LAW, TIE MINISTER OF IAW, JUSTICE AND 41n .JUSrICE AND CDMPAN! CCJ4PM1Y AFFAIRS A.'ID MINISTER OF J.52/5 A.!PAIRS AND MINIS'l'BR ~ 'ftWISPC&T ([1\. M. mA.'tB! OF SURP'ACE 'DWISl'CRr 'IRAI) (.au II. 'l.'IIMa1 1UlAt) 13&J.4/22(Col.4) 1,67,680 157/4 Y- (a) Yes 157/6 (a) lind (el (b) and (el 178/7 (bl (d) (fran be!0Ii) 183/2 3855 3755 196/16 (a) to (d) (a)(b) ~d (d) (fran below) 237/2 (e) Cd) (frau be!0Ii) 32:/16 (e) Ce) CONTENTS (Twelfth SerIN, Vol. III, Second Session, 1920 (Saa) No. 20, Monday, July 13, 199B1As.dhs 22, 1920 (Sea) SUBJECT • COLLIMN WRITTEN ANSWERS TO QUESTIONS 'Starred Questions Nos. 362-381. .... 3: ..... Unstarred Questions Nos. 3643-3872. 46-337 PAPERS LAID ON THE TABLE ............................................. -

Junior Assistant-2015-16 (Final)
NAME OF SHORT LISTED CANDIDATES FOR WRITTEN EXAMINATION OF JUNIOR ASSISTANT 2015-16 OF DEPUTTY COMMISSIONER'S OFFICE SONITPUR TEZPUR ROLL NAME ADDRESS DATE OF QUALIFIC OTHER CASTE NO. BIRTH ATION QUALIFICATI ON 1 Ajitav Choudhury S/O- Lt Akash Pratim 7/1/1991 HSLC Diploma in General Choudhury, Computer Vill- North Ghy, Rongmahal, PS- Changsari, PO- Rangmahal, Ghy-781030 2 Anjana Devi W/O- Sri Dhiraj 12/31/1986 BA PGDCA General Upadhyaya, (1Year) PO- Samardalani, PS- Sootea, Sonitpur 3 Amrita Talukdar D/O- Sri Binod Talukdar, 2/10/1983 HSLC DCA General Vill- Mazgaon, PO/PS- Tezpur 4 Anuradha Kalita W/o- Sri Khanindra 5/31/1979 BA 1 year computer General Saikia, Vill- Rangamati, Dist- Darrang, Pin-785429 5 Ananta Talukdar S/O- Barun talukdar, 11/30/1978 HSLC Library General Vill- Singimari, Technician/ PO- Kocharigaon, Typing English Pin-784117, Sonitpur Computer CCA 6 Atul Pachani S/O- Mr. KhaGeneral 4/30/1994 HSSLC ADCA ,Web - Pachani, Page Design Vill- Dubia, PO- Bishnupur, Dist- Dhemaji, Pin-787057 7 Amrit Das S/O- Sri Chenaram Das, 11/1/1977 HSSLC Computer SC Vill- Raidongia, Diploma PO-Aibheti, PS- Japori, Pin- 782002, Dist- Nagaon 8 Archana kalita D/O- Mr Nripen kalita, 12/27/1990 HSSLC PGDCA(1 Year) General Vill-Mathurapur(South), PO/PS- Tihu, Dist- Nalbari, Pin-781371 9 Ankana Hazarika D/O- Purnakanta 7/12/1993 BA PGDCA - Hazarika, Vill- Sonarigaon, PO- Silghat, Dist- Nagaon, 10 Arunabh Saikia S/O- Sri Amiya Kr Saikia, 6/6/1987 HSSLC Diploma in basic - Vill- Darrang College Computer Road, PO/PS- Tezpur 11 Ashim Das S/O- Sri Uma Kanta Das, 1/25/1992 BA Diploma in - Vill- Amloga, Computer PO- Uttar Amloga, PS- Chariduar. -

महहला एवं बाल हवकास Women and Child Development पूवोत्तर क्षेत्
महहला एवं बाल हवकास Women and Child Development पूवोत्तर क्षेत्र के राज्यों के हलए प्रावधान पर टिप्पण Note on provision for States of North Eastern Area वषष 2014-15 के बजि प्राक्कलन में उप-शीषष के सामने In the Budget Estimates for 2014-15 the Plan outlay figures indicated against the sub-heads are exclusive उल्हलहखत योजना पटरव्यय के आंकडों में पूवोत्तर क्षेत्र के of North Eastern Area provision as 10% of Central हलए केन्द्रीय योजना प्रावधान का दस प्रहतशत शाहमल नहीं Plan provision, i.e Rs. 2110.00 crores has been kept है अथाषत पूवोत्तर क्षेत्र के लाभाथष मुख्य शीषष 2552 के separately under Major Head 2552 for the benefit of अंतगषत 2110 करोड 셁पए अलग रखे गए हैं । वषष के the North Eastern Areas. During the course of the year, funds will be re-appropriated from this provision दौरान पूवोत्तर क्षेत्र हेतु हवहभन्न स्कीमों के अंतगषत to meet the requirement under various schemes for आवश्यकता䴂 की पूर्तत हेतु इस प्रावधान में से राहश का North Eastern Areas. पुनर्तवहनयोजन ककया जाएगा । हवषय सूची TABLE OF CONTENTS महहलाएवं बाल हवकास मंत्रालय MINISTRY OF WOMEN AND CHILD DEVELOPMENT Page No. 1.अनुदान मांगों का ब्यौरा मांग संख्या 99 Detailed Demands for Grants / Demand No. 99 1-139 महहला एवं बाल हवकास मंत्रालय Ministry of Women and Child Development 2. शीषषवार ब्यौरेवार व्यवस् था दशाषनेवाला हववरण Statement showing detailed object Head-wise provision 140-141 3.पटरहशष्ि-IX-क-कमषचाटरयों की अनुमाहनत संख्या तथा तत्संबंधी प्रावधान Appendix-IX-A- Estimated strength of Establishment and provision therefor 142-146 4.पटरहशष्ि-IX-ख- केंद्रीययोजनामेंबासहायताप्राप्त पटरयोजना䴂परव्ययकेहलएपटरयोजना- Apendix –IX-B Project-wise provision for expenditure on Externally-Aided projects 147 वारप्रावधान in the Central Plan 5. -
DETAILS of the Phd SCHOLARS (Under 2009 and 2017 Regulations)
ASSAM UNIVERSITY, SILCHAR DETAILS OF THE PhD SCHOLARS (Under 2009 and 2017 Regulations) Dept: Agricultural Engineering Sl. School Department Name & Address of Name of the Mode of Registration / Research Topic Tentative date Availing Funding Agency No. the PhD Scholar Supervisor & pursuing Enrolment of completion of Fellowship & of Fellowship with e-mail ID and Co-Supervisor PhD (Full Number & PhD Course Name (Yes/No) Contact No. & (if any) Time/Part Date of Adhaar No/ Time Registration / Voter Card No./ Admission Driving Licence No./Passport No. 1. Triguna Sen Agricultural Rishi Kumar, Dr. Laxmi Part Time Ph.D./2269/13 Microbial removal of heavy 2019 No - School of Engineering e-mail Narayan Sethi Dt: 26.02.13 metals from aquatic Technology ID:rishisingh131@ Dr. Sudipto environment using gmail.com, Phone Saqrkar bioreactors. No.: 9717346600 Aadhaar No.- 583732874806 2. Triguna Sen Agricultural Kamakshi Prasad Dr. Laxmi Part Time Ph.D./2362/13 Integrating water 2019 No - School of Engineering Padhy, S/o: Sri Narayan Sethi Dt: 12.09.13 harvesting and hydraulic Technology Raghunath Prasad ram fed micro-irrigation Padhy, AT/P O: system for crop planning in Medical Bank hilly terrain Colony-II, Berhampur, Distt.: Ganjam, Odisha, PIN: 760004, e- mail ID: kamakshipadhy@y ahoo.com, kppadhy77@gmail. com, Phone No.: 09438055369 3. Triguna Sen Agricultural Gajendra Prasad, Dr. Sudipto Part Time PhD/SOT/10/1 Design and development of No - School of Engineering Department of Sarkar 6 solar biomass hybrid drying Technology Agricultural Dr. Laxmi Dtd. system for fruits and Engineering, Narayan Sethi 13/04/2016 vegetables Triguna Sen School of Technology, Assam University, Silchar-788011, e-mail ID: iitkgp.gajendra@g mail.com, Phone No.: 8811083834 Adhaar No.- 848895343201, 7488218615 4. -
Genetic Diversity and Population Structure of Monopterus Spp. Of
International Journal of Fisheries and Aquatic Studies 2017; 5(5): 457-464 E-ISSN: 2347-5129 P-ISSN: 2394-0506 (ICV-Poland) Impact Value: 5.62 Genetic Diversity and Population Structure of (GIF) Impact Factor: 0.549 IJFAS 2017; 5(5): 457-464 Monopterus spp. of Assam and Manipur states of © 2017 IJFAS www.fisheriesjournal.com Northeast India using Microsatellite Markers Received: 27-07-2017 Accepted: 28-08-2017 Papari Devi, MA Laskar, Chittaranjan Baruah and DK Sharma Papari Devi Department of Zoology, Kaliabor College, Kuwarital, Abstract Assam, India Eleven microsatellite markers were used to sort out genetic diversity and population differentiation of Monopterus species complex of Northeast India. Samples of morphologically identified M. albus (50 MA Laskar nos.) and M. cuchia (180 nos.) were collected from Manipur and Assam, were allocated into four Department of Biotechnology, populations based on the geographical proximity of the water bodies. Polymorphism Information Content St. Anthony's College, Shillong, (PIC) across the eleven microsatellite loci ranged from 0.588 to 0.852. The mean single locus estimate of Meghalaya, India Wright’s inbreeding indices FIT and FIS were 0.132 and 0.177 and mean single locus estimate of Wright’s fixation index FST was 0.052. The average numbers of observed alleles (NA) and effective alleles (NAe) Chittaranjan Baruah across 11 microsatellite loci were 9.73 and 6.33 respectively, suggestive of moderate heterozygosity. The Department of Zoology, Darrang mean expected heterozygosity within and among individuals of M. albus was 0.573 and 0.709 College, Tezpur, Assam, India respectively while these values for M. -

PLICATE Hrtdisti NAGAON ASSAM 782137ANK OE NDIA HTHT
PLICATE KALIABOR P.O KUWARITOL HRTDiSTI NAGAON ASSAM 782137ANK OE NDIA npt a Negatiable Document Tel: 276616 ALIABOR COL OF EDUCATION ILL: KUWARITAL TERM DEPOSIT ADVICE PO: KUWARITAL 06/08/2019 DIST: NAGAON (In lieu of Term Deposit Receipt) Nagaon HTHT/Nomination it5/Registered l3Tip/Not Registered HT5/Date: E/ TETET, Dear SirMadam Ya3I,EA IR aEI We have pleasure in confiming details of the folowing amount held in deposit with us. PleLA3GE AAa&bRGMBSh all corespondence. 8ha796T4ot Banking with us. AND REGtONAL DIRECTR AT/Name: RC NTE AT/CIF No. Bhubonekw an A d T/PAN No. SINGLE STD-GEN PUB-OTi-5Y-10Y-INR Mode of Operation: Scheme H /Alc No. aTsTerm: Interest @ Pripal Amt. Value Date Maturity Date 35322150882 7 % 4.00000.00 311 2015 3442020- HMd( Vaue00 HaqT/Yours faithfuly A Printed 2 Times Terg areriAuthorieed Signatory 99 grKALIABOR OF INDAINDIA This is riot a Negotiable Document XDISTT, NAGAONPQKUWARITOLBANK ASSAN Y6223ANK Tel: 276616 HTaT T KALIABOR COL OF EDUCATION TERM DEPOSIT ADVICE viLL: KUVWARITAL 06/08/2019 PO: KUWARITAL (in lieu of Term Deposit Receipt) DIST: NAGAON Nagaon HTT/Nomination tiT Registered 3Tipa Not Registered TH Date T4/ TE441, Dear SirMadam i sr1 zar zaa arT f re We have Dleasure n coniming detals of the folowing amount held in deposit with us. PlaneLABOR tororeDuOAtHONN all corespondence. tssd7Devdor Banking with us. AN R EhtoNAL RECTDN, ERG Name: NCTE Bhsbanesed CIF No. d PAN No. SINGLE STD-GEN PUB-OTH-5Y-10Y-INR Mode of Operation Scheme ATET t art RT H /Alc No.: rareJerm : Interest Prinsal Amt Vatue Date Maturity -

Page 1 of 7 in the COURT of the MEMBER, MOTOR ACCIDENT
IN THE COURT OF THE MEMBER, MOTOR ACCIDENT CLAIMS TRIBUNAL SONITPUR AT TEZPUR Present : Smti. M. Nandi., Member, Motor Accident Claims Tribunal, Sonitpur, Tezpur MAC Case No. 279 of 2012(P) 1. Smti. Niru Bharali, W/O Late Karun Bharali Vill – Murhadol, Solaguri P.O. & P.S. – Jamuguri Dist – Sonitpur, Assam -Versus- 1. New India Assurance Co. Ltd. Tezpur Branch, Tezpur Dist – Sonitpur, Assam 2. Md. Azibul Hussain, S/O- Late Ratigul Hussain. Vill Kuwarital Kaliabor, Nagaon, P.O & P.S. Kaliabar Dist –Nagaon, Assam 3. Md. Saiful Islam S/O Md. Gulam Hussain, R/O No.1 Bhurbandha P.O. Lowkhowa P.S. Rupahihat. Dist. Nagaon, Assam. …………………….. opposite parties. ADVOCATES APPEARED For the claimant : -Sri S. Khan, Advocate. For the O.P. No.1 :- Sri P. Sethi, Advocate. For the O.P. NO. 2 & 3 :- Ex-parte. Date of Argument :-22-11-2017. Date of Judgment :-30-11-2017. Page 1 of 7 J U D G M E N T The claimant Smti. Niru Bharali has filed an application u/s 166 of MV Act, 1988, claiming compensation on account of damage of her vehicle bearing No. AS-12G-3135 in a motor vehicle accident. 1. The brief fact of the case is that on 11-07-12 at about 4-00 A.M. the vehicle bearing No. AS-12G/3135 (Bolero) was going from Tezpur towards Kaliabor . When the said vehicle reached near Kaliabor Bridge at about 4-30 A.M. another vehicle bearing No. AS-12C/2548 ( 207 DI Tata Truck) was coming from opposite direction with loaded timbers and due to rash and negligent driving, the driver of the said vehicle could not control himself and hit the vehicle of the claimant bearing No. -
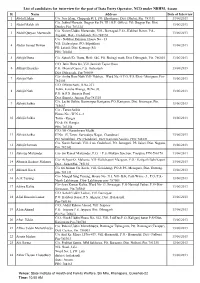
Data Entry Operator-1
List of candidates for interview for the post of Data Entry Operator, NCD under NRHM, Assam Sl. Name Address Date of Interview 1 Abdul Hakim C/o. Nur Islam, Chapgarh Pt. I, PO. Khudimari, Dist. Dhubri, Pin. 783331 13/06/2013 C/o. Jobbul Hussain, Jhagrar Par Pt. III ( G.P. Office), PO. Jhagrar Par, Dist. 2 Abdul Malek Ali 13/06/2013 Dhubri, Pin. 783324 C/o Nizam Uddin Mazumder, Vill.- Barnagad, P.O.- Kalibari Bazar, P.S.- 3 Abdul Quiyam Mazumder 13/06/2013 Algapur, Dist.- Hailakandi, Pin-788150 C/o - Nabibur Rahman, House No - 33 Vill: Hedayetpur, PO: Silpukhuri 4 Abdus Samad Dewan 13/06/2013 PS: Latasil, Dist: Kamrup (M) PIN: 781003 5 Abhijit Dutta C/o. Satosh Kr. Dutta, HnO. 646, PO. Boiragi math, Dist. Dibrugarh, Pin. 786003 13/06/2013 C/O. Jatin Hazarika, Vill. Jamirah Tepor Gaon 6 Abhijit Hazarika P.O. Dhamal Gaon, P.S. Barbaruah 13/06/2013 Dist. Dibrugarh, Pin-786004 C/o- Ambu Ram Nath Vill- Nabheti, Ward No.-5 P.O./P.S./Dist.- Morigaon, Pin- 7 Abhijit Nath 13/06/2013 782105 C/O. Dhiren Nath, H.No.231 Town. Kalaha Bhanga, W/No.03, 8 Abhijit Nath 13/06/2013 P.O. & P.S. Barpeta Road Dist. Barpeta, Assam, Pin-781315 C/o. Lachit Saikia, Konwarpur Karigaon, PO. Karigaon, Dist. Sivasagar, Pin. 9 Abhijit Saikia 13/06/2013 785667 C/o - Tarun Saikia House No - W.N.o -3 10 Abhijit Saikia Town - Rangia 13/06/2013 PO & PS: Rangia PIN: 781354 C/O: Mr Ghanashyam Medhi 11 Abhijit Saikia H No: 15, Town: Sarvodaya Nagar, Chandmari 13/06/2013 PO: Silpukhuri, PS: Chandmari, Dist: Kamrup(Assam), PIN: 781021 C/o. -

TOWARDS ENDOWMENT FUND and /OR 316:Era
LO SBI State Bank of India CM/19/ 3s Dt:06/08/2019 The Regional Director, Eastern Regional Committee National Council for Teacher Education 15, Neelakantha Nagar, Nayapalli Bhubaneswar, Odisha 751012 Dear Sir, CONVERSION/RENEWAL OF FIXED DERPOSIT RECEIPT(S) TOWARDS ENDOWMENT FUND AND /OR RESERVE FUND IN THE JOINT NAME OF THE SOCIETY/TRUST/INSTITUTIQN AND REGIONAL DIRECTOR ERC, NCTE, BHUBANESWAR(Joint operation Mode: "AND') the fixed Receipt No On receipt of authorization from the Eastern Regional Committee, NCTE, Deposit 35535724562 dated 29/01/2016 for Rs. 5.00 lakhs (Rupees five lakhs only) for a period of 5 years from EDUCATION with the name of 29/01/2016 To 29/01/2021 the joint name of KALIABOR COLLEGE OF Society/Trust/Institution and Regional Director, ERC, NCTE, Bhubaneswr has been converted/renewed. Further, the Fixed Deposit Receipt No 355357458 ted 29/01/2016 for Rs. 3.00 lakhs (Rupees three lakhs only) and Fixed Deposit Receipt No 353. 2 dated 03/11/2015 for Rs. 4.00 lakhs (Rupees four lakhs only)for a period of 5 years from 29/01/-16 to 29/01/2021 & 03/11/2015 to 03/11/2020 respectively, in the joint name of KALIABOR COLLEGE OF EDUCATION and Regional Director, ERC, NCTE, Bhubaneswar has been converted/renewed. The application for Fixed Deposit Receiptis) has been signed by Secretary and Preside nt of Kaliabor College of Education. The Fixed Deposit Receipt(s) converted/renewed have been handed over, to the Secretary, Kaliabor college of Education of the Society/Trust/Institution who sends the same to ERC, NCTE Office. -

As Per Advertisement No. Janasanyog/2665/15, Dtd.15/06/2015 the List Of
As per Advertisement No. Janasanyog/2665/15, dtd.15/06/2015 the list of candidate who have already applied for the post of Ward Boy/Ward Girl has been listed bellow and they need not apply again: Name & Address Sl. No. SURJYA BHANU, ABDUL SALAM, VILL- 1 BALARCHAR, P.O- KIRTANPARA, DIST- BANGAIGAON, ASSAM, PIN-780084 MITALI DEVI, LT. HARENDRA NATH SARMA, 2 CHRISTANBASTI UDAYACHALSAMMONNAY PATH, H. NO- 15, P.O- DISPUR, GHY-5 SHRI HIMANGSHU NATH, C/O- SHRI SUKLESHWAR NATH, P.O- 3 BAMUNIMAIDAN, DIST- KAMRUP,ASSAM, GHY-3, PIYALI PHUKAN NAGAR SRI MANAB DAS, C/O- SRI NAGESWAR DAS, 4 VILL- MAJPARA, P.O- CHAYGAON, DIST- KAMRUP,ASSAM, PIN-781124 SRI KISHOR MEDHI, C/O- PRAHLAD MEDHI, 5 VILL+P.O- KHAGROBARI, DIST- BAKSA, ASSAM, PIN-781344 SUSMITA DEVI DEKA, C/O- BISHNU SARMA, PIYALI PHUKAN PATH, P.O- 6 BAMUNIMAIDAN, DIST- KAMRUP, ASSAM, GHY-3 Page 1 of 926 SRI SHANJIT KAIBARTA, C/O- ABHI RAM KAIBARTA, VILL- GARGARA, P.O- 7 SIKARHATI, DIST- KAMRUP,ASSAM, PIN- 781125 MONAJ NAG, C/O- MANIK CH. NAG, 8 ADABARI TINIALI G.R.P. RESERVE LANE BARIPARA, PIN-781012 DIPANKAR DAS, C/O- ANIL DAS, PANDU TEMPLE GHAT, NEAR QR. NO-9/B, P.O- 9 PANDU REST CAMO, DIST- KAMRUP, ASSAM,PIN-781012 ASHRAFUL HAQUE, C/O- LT. SAIFUDDIN 10 AHMED, VILL+P.O- KALITAKUCHI, DIST- KAMRUP,ASSAM, PIN-781102 RATUL CHANDRA BARO, C/O- LT. CHANA RAM BARO, VILL-JAJIKANA, P.O- 11 MODHUKUCHI, DIST- KAMRUP,ASSAM, PIN- 781354 ANJUMA BEGUM, C/O- ATOWAR RAHMAN, 12 VILL- KACHARIPETY PT-II, P.O- AMBARI, PIN-783384 SANJIT BISWAS, C/O- LT. -

Ngos REGISTERED UNDER SOCIETIES REGISTRATION ACT XXI of 1860 in the STATE of ASSAM (1957 to 1980)
NGOs REGISTERED UNDER SOCIETIES REGISTRATION ACT XXI OF 1860 IN THE STATE OF ASSAM (1957 To 1980) Name of the NGO registered under S.R. Act XXI of 1860 Address Registration Year/Date of District No. Registration BAPUJI PATHAGAR 19 1957 BARPETA BARPETA DAKSHIN BANG SEVA SANGHA 34 1957 DAKSHIN BANG BARPETA NITYA NANDA GRAM UNNAYAN WOMEN WELFARE CENTRE 37 1957 NITYANANDA BARPETA BARPETA BUS SYNDICATE 38 1957 P.O- BARPETA BARPETA PRAGATI SANGHA 44 1957 P.O- SARUPETA BARPETA SANATAN GANA SAKTI SAMITTEE 47 1957 BARPETA BARPETA THE TENNIS CLUB, SILCHAR 10 1957 P.O- SILCHAR CACHAR GITARTHI SAMAJ BALIGAON 14 1957 BALIGAON DARRANG CHAMUKHA ANCHALIK MAHILA-O-SISHU KALYAN SAMITY 1887 1957 VILL- CHAMUKHA P.O- CHENGELIAJHAN DARRANG MILAN SANGHA 30 1957 DIBRUGARH DIBRUGARH THE GOALPARA MOTORS VEHICLES ASSOCIATION 46 1957 P.O- GOALPARA GOALPARA SINDHI ASSOCIATION 17 1957 GUWAHATI KAMRUP (M) SRI RAM KRISHNA SEVA SAMITTEE 18 1957 GUWAHATI KAMRUP (M) REHABARI UNNAYAN SAMITY 20 1957 GUWAHATI KAMRUP (M) T.R PHOOKAN SMRITI BHAWAN RANGA MANCHA 21 1957 GUWAHATI KAMRUP (M) GRAM SEVAK SANGHA 22 1957 GUWAHATI KAMRUP (M) Page 1 of 280 ASSAM PHARMATEUTICAL ASSOCIATION 28 1957 GUWAHATI KAMRUP (M) ASSAM SHISHU KALYAN SADAN 32 1957 GUWAHATI KAMRUP (M) THE ASSAM STATE COUNCIL FOR CHILD WELFARE 35 1957 GUWAHATI KAMRUP (M) THE TRUST ASSOCIATION OF THE ASSEMBLY OF THE PRESBYTERIAN 40 1957 GUWAHATI KAMRUP (M) CHURCH IN ASSAM DISPUR BORO BIHAR SAMITTEE 43 1957 GUWAHATI DISPUR KAMRUP (M) FANCY BAZAR MUNICIPAL MARKET DEALERS ASSOCIATION 45 1957 GUWAHATI FANCY BAZAR KAMRUP (M)