Dna Barcoding of Chlorarachniophytes Using Nucleomorph Its Sequences1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Glycolytic Reactions in the Mitochondrion?
Broad Distribution of TPI-GAPDH Fusion Proteins among Eukaryotes: Evidence for Glycolytic Reactions in the Mitochondrion? Takuro Nakayama1, Ken-ichiro Ishida2, John M. Archibald1* 1 Department of Biochemistry & Molecular Biology, Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity, Dalhousie University, Halifax, Nova Scotia, Canada, 2 Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Ibaraki, Japan Abstract Glycolysis is a central metabolic pathway in eukaryotic and prokaryotic cells. In eukaryotes, the textbook view is that glycolysis occurs in the cytosol. However, fusion proteins comprised of two glycolytic enzymes, triosephosphate isomerase (TPI) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were found in members of the stramenopiles (diatoms and oomycetes) and shown to possess amino-terminal mitochondrial targeting signals. Here we show that mitochondrial TPI- GAPDH fusion protein genes are widely spread across the known diversity of stramenopiles, including non-photosynthetic species (Bicosoeca sp. and Blastocystis hominis). We also show that TPI-GAPDH fusion genes exist in three cercozoan taxa (Paulinella chromatophora, Thaumatomastix sp. and Mataza hastifera) and an apusozoan protist, Thecamonas trahens. Interestingly, subcellular localization predictions for other glycolytic enzymes in stramenopiles and a cercozoan show that a significant fraction of the glycolytic enzymes in these species have mitochondrial-targeted isoforms. These results suggest that part -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

Chromerid Genomes Reveal the Evolutionary Path From
RESEARCH ARTICLE elifesciences.org Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites Yong H Woo1*, Hifzur Ansari1,ThomasDOtto2, Christen M Klinger3†, Martin Kolisko4†, Jan Michalek´ 5,6†, Alka Saxena1†‡, Dhanasekaran Shanmugam7†, Annageldi Tayyrov1†, Alaguraj Veluchamy8†§, Shahjahan Ali9¶,AxelBernal10,JavierdelCampo4, Jaromır´ Cihla´ rˇ5,6, Pavel Flegontov5,11, Sebastian G Gornik12,EvaHajduskovˇ a´ 5, AlesHorˇ ak´ 5,6,JanJanouskovecˇ 4, Nicholas J Katris12,FredDMast13,DiegoMiranda- Saavedra14,15, Tobias Mourier16, Raeece Naeem1,MridulNair1, Aswini K Panigrahi9, Neil D Rawlings17, Eriko Padron-Regalado1, Abhinay Ramaprasad1, Nadira Samad12, AlesTomˇ calaˇ 5,6, Jon Wilkes18,DanielENeafsey19, Christian Doerig20, Chris Bowler8, 4 10 3 21,22 *For correspondence: yong. Patrick J Keeling , David S Roos ,JoelBDacks, Thomas J Templeton , 12,23 5,6,24 5,6,25 1 [email protected] (YHW); arnab. Ross F Waller , Julius Lukesˇ , Miroslav Obornık´ ,ArnabPain* [email protected] (AP) 1Pathogen Genomics Laboratory, Biological and Environmental Sciences and Engineering † These authors contributed Division, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia; equally to this work 2Parasite Genomics, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Present address: ‡Vaccine and Cambridge, United Kingdom; 3Department of Cell Biology, University of Alberta, Infectious Disease Division, Fred Edmonton, Canada; 4Canadian Institute for Advanced Research, Department of Botany, -

The Apicoplast: a Review of the Derived Plastid of Apicomplexan Parasites
Curr. Issues Mol. Biol. 7: 57-80. Online journalThe Apicoplastat www.cimb.org 57 The Apicoplast: A Review of the Derived Plastid of Apicomplexan Parasites Ross F. Waller1 and Geoffrey I. McFadden2,* way to apicoplast discovery with studies of extra- chromosomal DNAs recovered from isopycnic density 1Botany, University of British Columbia, 3529-6270 gradient fractionation of total Plasmodium DNA. This University Boulevard, Vancouver, BC, V6T 1Z4, Canada group recovered two DNA forms; one a 6kb tandemly 2Plant Cell Biology Research Centre, Botany, University repeated element that was later identifed as the of Melbourne, 3010, Australia mitochondrial genome, and a second, 35kb circle that was supposed to represent the DNA circles previously observed by microscopists (Wilson et al., 1996b; Wilson Abstract and Williamson, 1997). This molecule was also thought The apicoplast is a plastid organelle, homologous to to be mitochondrial DNA, and early sequence data of chloroplasts of plants, that is found in apicomplexan eubacterial-like rRNA genes supported this organellar parasites such as the causative agents of Malaria conclusion. However, as the sequencing effort continued Plasmodium spp. It occurs throughout the Apicomplexa a new conclusion, that was originally embraced with and is an ancient feature of this group acquired by the some awkwardness (“Have malaria parasites three process of endosymbiosis. Like plant chloroplasts, genomes?”, Wilson et al., 1991), began to emerge. apicoplasts are semi-autonomous with their own genome Gradually, evermore convincing character traits of a and expression machinery. In addition, apicoplasts import plastid genome were uncovered, and strong parallels numerous proteins encoded by nuclear genes. These with plastid genomes from non-photosynthetic plants nuclear genes largely derive from the endosymbiont (Epifagus virginiana) and algae (Astasia longa) became through a process of intracellular gene relocation. -

Testing the Effect of in Planta RNA Silencing on Plasmodiophorabrassicae Infection
Testing the effect of in planta RNA silencing on Plasmodiophorabrassicae infection A thesis submitted in partial fulfilment of the requirements for the Degree of Doctor of Philosophy at Lincoln University S. R. Bulman 2006 ii Contents Abstract. ...................................................................................................., ................ v Acknowledgements .................................................................................................. vii List of Tables ........................................................................................................... viii List of Figures ........................................................................................................... ix CHAPTER 1. Literature Review ................................................................................ 1 Plasmodiophora brassicae ............................................................................................. 1 RNA silencing .................................................................................................................. 5 Plant defence by RNA silencing .................................................................................... 8 RNA silencing versus P. brassicae ............................................................................. 10 Molecular biology of P. brassicae ................................................................................ 10 Choice of cDNA cloning strategy in P. brassicae ...................................................... -

Plasmodiophora Brassicae
Bi et al. Phytopathology Research (2019) 1:12 https://doi.org/10.1186/s42483-019-0018-6 Phytopathology Research RESEARCH Open Access Comparative genomics reveals the unique evolutionary status of Plasmodiophora brassicae and the essential role of GPCR signaling pathways Kai Bi1,2, Tao Chen2, Zhangchao He1,2, Zhixiao Gao1,2, Ying Zhao1,2, Huiquan Liu3, Yanping Fu2, Jiatao Xie1,2, Jiasen Cheng1,2 and Daohong Jiang1,2* Abstract Plasmodiophora brassicae is an important biotrophic eukaryotic plant pathogen and a member of the rhizarian protists. This biotrophic pathogen causes clubroot in cruciferous plants via novel intracellular mechanisms that are markedly different from those of other biotrophic organisms. To date, genomes from six single spore isolates of P. brassicae have been sequenced. An accurate description of the evolutionary status of this biotrophic protist, however, remains lacking. Here, we determined the draft genome of the P. brassicae ZJ-1 strain. A total of 10,951 protein-coding genes were identified from a 24.1 Mb genome sequence. We applied a comparative genomics approach to prove the Rhizaria supergroup is an independent branch in the eukaryotic evolutionary tree. We also found that the GPCR signaling pathway, the versatile signal transduction to multiple intracellular signaling cascades in response to extracellular signals in eukaryotes, is significantly enriched in P. brassicae-expanded and P. brassicae-specific gene sets. Additionally, treatment with a GPCR inhibitor relieved the symptoms of clubroot and significantly suppressed the development of plasmodia. Our findings suggest that GPCR signal transduction pathways play important roles in the growth, development, and pathogenicity of P. brassicae. -

Provirophages in the Bigelowiella Genome Bear Testimony to Past Encounters with Giant Viruses
Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses Guillaume Blanca,1,2, Lucie Gallot-Lavalléea, and Florian Maumusb,1,2 aLaboratoire Information Génomique et Structurale, UMR7256 (Institut de Microbiologie de la Méditerranée FR3479) CNRS, Aix-Marseille Université, 13288 Marseille cedex 9, France; and bINRA, UR1164 Unité de Recherche Génomique-Info, Institut National de la Recherche Agronomique de Versailles-Grignon, 78026 Versailles, France Edited by Peter Palese, Icahn School of Medicine at Mount Sinai, New York, NY, and approved July 24, 2015 (received for review April 1, 2015) Virophages are recently discovered double-stranded DNA virus satel- cysteine protease (PRO), and zinc-ribbon domain (ZnR) as well as lites that prey on giant viruses (nucleocytoplasmic large DNA viruses; major and minor capsid proteins (MCPs and mCPs, respectively) NCLDVs), which are themselves parasites of unicellular eukaryotes. (12). In addition, genes encoding two different families of integrases This coupled parasitism can result in the indirect control of eukaryotic have been identified in several virophages: A putative rve integrase cell mortality by virophages. However, the details of such tripartite was found in Mavirus and ALM (8, 10), whereas Sputnik encodes a ∼ relationships remain largely unexplored. We have discovered 300 putative tyrosine integrase (1). Among virophage genes, only PRO, predicted genes of putative virophage origin in the nuclear genome ATPase, MCP, and mCP support the monophyly of virophages, of the unicellular alga Bigelowiella natans. Physical clustering of these whereas the remaining gene complement shows complex phyloge- genes indicates that virophage genomes are integrated into the B. natans genome. Virophage inserts show high levels of similarity nies suggestive of gene replacement (12). -

Phylogenomics Supports the Monophyly of the Cercozoa T ⁎ Nicholas A.T
Molecular Phylogenetics and Evolution 130 (2019) 416–423 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogenomics supports the monophyly of the Cercozoa T ⁎ Nicholas A.T. Irwina, , Denis V. Tikhonenkova,b, Elisabeth Hehenbergera,1, Alexander P. Mylnikovb, Fabien Burkia,2, Patrick J. Keelinga a Department of Botany, University of British Columbia, Vancouver V6T 1Z4, British Columbia, Canada b Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok 152742, Russia ARTICLE INFO ABSTRACT Keywords: The phylum Cercozoa consists of a diverse assemblage of amoeboid and flagellated protists that forms a major Cercozoa component of the supergroup, Rhizaria. However, despite its size and ubiquity, the phylogeny of the Cercozoa Rhizaria remains unclear as morphological variability between cercozoan species and ambiguity in molecular analyses, Phylogeny including phylogenomic approaches, have produced ambiguous results and raised doubts about the monophyly Phylogenomics of the group. Here we sought to resolve these ambiguities using a 161-gene phylogenetic dataset with data from Single-cell transcriptomics newly available genomes and deeply sequenced transcriptomes, including three new transcriptomes from Aurigamonas solis, Abollifer prolabens, and a novel species, Lapot gusevi n. gen. n. sp. Our phylogenomic analysis strongly supported a monophyletic Cercozoa, and approximately-unbiased tests rejected the paraphyletic topologies observed in previous studies. The transcriptome of L. gusevi represents the first transcriptomic data from the large and recently characterized Aquavolonidae-Treumulida-'Novel Clade 12′ group, and phyloge- nomics supported its position as sister to the cercozoan subphylum, Endomyxa. These results provide insights into the phylogeny of the Cercozoa and the Rhizaria as a whole. -

New Phylogenomic Analysis of the Enigmatic Phylum Telonemia Further Resolves the Eukaryote Tree of Life
bioRxiv preprint doi: https://doi.org/10.1101/403329; this version posted August 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote tree of life Jürgen F. H. Strassert1, Mahwash Jamy1, Alexander P. Mylnikov2, Denis V. Tikhonenkov2, Fabien Burki1,* 1Department of Organismal Biology, Program in Systematic Biology, Uppsala University, Uppsala, Sweden 2Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, Yaroslavl Region, Russia *Corresponding author: E-mail: [email protected] Keywords: TSAR, Telonemia, phylogenomics, eukaryotes, tree of life, protists bioRxiv preprint doi: https://doi.org/10.1101/403329; this version posted August 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract The broad-scale tree of eukaryotes is constantly improving, but the evolutionary origin of several major groups remains unknown. Resolving the phylogenetic position of these ‘orphan’ groups is important, especially those that originated early in evolution, because they represent missing evolutionary links between established groups. Telonemia is one such orphan taxon for which little is known. The group is composed of molecularly diverse biflagellated protists, often prevalent although not abundant in aquatic environments. -

Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella Natans
J. Eukaryot. Microbiol., 51(5), 2004 pp. 529±535 q 2004 by the Society of Protozoologists Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella natans MATTHEW B. ROGERS,a JOHN M. ARCHIBALD,a,1 MATTHEW A. FIELD,a CATHERINE LI,b BORIS STRIEPENb and PATRICK J. KEELINGa aCanadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC, V6T 1Z4, Canada, and bCenter for Tropical & Emerging Global Diseases & Department of Cellular Biology, University of Georgia, 724 Biological Sciences Building, Athens, Georgia 30602, USA ABSTRACT. Chlorarachniophytes are marine amoebo¯agellate protists that have acquired their plastid (chloroplast) through secondary endosymbiosis with a green alga. Like other algae, most of the proteins necessary for plastid function are encoded in the nuclear genome of the secondary host. These proteins are targeted to the organelle using a bipartite leader sequence consisting of a signal peptide (allowing entry in to the endomembrane system) and a chloroplast transit peptide (for transport across the chloroplast envelope mem- branes). We have examined the leader sequences from 45 full-length predicted plastid-targeted proteins from the chlorarachniophyte Bigelowiella natans with the goal of understanding important features of these sequences and possible conserved motifs. The chemical characteristics of these sequences were compared with a set of 10 B. natans endomembrane-targeted proteins and 38 cytosolic or nuclear proteins, which show that the signal peptides are similar to those of most other eukaryotes, while the transit peptides differ from those of other algae in some characteristics. Consistent with this, the leader sequence from one B. -
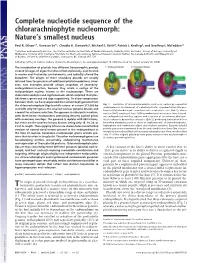
Complete Nucleotide Sequence of the Chlorarachniophyte Nucleomorph: Nature’S Smallest Nucleus
Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus Paul R. Gilson*†, Vanessa Su†‡, Claudio H. Slamovits§, Michael E. Reith¶, Patrick J. Keeling§, and Geoffrey I. McFadden‡ʈ *Infection and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Parkville 3050, Australia; ‡School of Botany, University of Melbourne, Victoria 3010, Australia; ¶Institute for Marine Biosciences, National Research Council, Halifax, NS, Canada B3H 3Z1; and §Department of Botany, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Edited by Jeffrey D. Palmer, Indiana University, Bloomington, IN, and approved April 19, 2006 (received for review January 26, 2006) The introduction of plastids into different heterotrophic protists created lineages of algae that diversified explosively, proliferated in marine and freshwater environments, and radically altered the biosphere. The origins of these secondary plastids are usually inferred from the presence of additional plastid membranes. How- ever, two examples provide unique snapshots of secondary- endosymbiosis-in-action, because they retain a vestige of the endosymbiont nucleus known as the nucleomorph. These are chlorarachniophytes and cryptomonads, which acquired their plas- tids from a green and red alga respectively. To allow comparisons between them, we have sequenced the nucleomorph genome from the chlorarachniophyte Bigelowiella natans: at a mere 373,000 bp Fig. 1. Evolution of chlorarachniophytes such as B. natans by sequential endosymbioses. Enslavement of a photosynthetic, cyanobacterium-like pro- and with only 331 genes, the smallest nuclear genome known and karyote (Cb) introduces photosynthesis into a eukaryotic host (Euk 1), whose a model for extreme reduction. The genome is eukaryotic in nature, nucleus (Nu1) acquires at least 1,000 cyanobacterial genes over time. -

Living Organisms Author Their Read-Write Genomes in Evolution
biology Review Living Organisms Author Their Read-Write Genomes in Evolution James A. Shapiro ID Department of Biochemistry and Molecular Biology, University of Chicago GCIS W123B, 979 E. 57th Street, Chicago, IL 60637, USA; [email protected]; Tel.: +1-773-702-1625 Academic Editor: Andrés Moya Received: 23 August 2017; Accepted: 28 November 2017; Published: 6 December 2017 Abstract: Evolutionary variations generating phenotypic adaptations and novel taxa resulted from complex cellular activities altering genome content and expression: (i) Symbiogenetic cell mergers producing the mitochondrion-bearing ancestor of eukaryotes and chloroplast-bearing ancestors of photosynthetic eukaryotes; (ii) interspecific hybridizations and genome doublings generating new species and adaptive radiations of higher plants and animals; and, (iii) interspecific horizontal DNA transfer encoding virtually all of the cellular functions between organisms and their viruses in all domains of life. Consequently, assuming that evolutionary processes occur in isolated genomes of individual species has become an unrealistic abstraction. Adaptive variations also involved natural genetic engineering of mobile DNA elements to rewire regulatory networks. In the most highly evolved organisms, biological complexity scales with “non-coding” DNA content more closely than with protein-coding capacity. Coincidentally, we have learned how so-called “non-coding” RNAs that are rich in repetitive mobile DNA sequences are key regulators of complex phenotypes. Both biotic and abiotic ecological challenges serve as triggers for episodes of elevated genome change. The intersections of cell activities, biosphere interactions, horizontal DNA transfers, and non-random Read-Write genome modifications by natural genetic engineering provide a rich molecular and biological foundation for understanding how ecological disruptions can stimulate productive, often abrupt, evolutionary transformations.