Lessons Learned from the Dog Genome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Approximating Selective Sweeps
Approximating Selective Sweeps by Richard Durrett and Jason Schweinsberg Dept. of Math, Cornell U. Corresponding Author: Richard Durrett Dept. of Mathematics 523 Malott Hall Cornell University Ithaca NY 14853 Phone: 607-255-8282 FAX: 607-255-7149 email: [email protected] 1 ABSTRACT The fixation of advantageous mutations in a population has the effect of reducing varia- tion in the DNA sequence near that mutation. Kaplan, Hudson, and Langley (1989) used a three-phase simulation model to study the effect of selective sweeps on genealogies. However, most subsequent work has simplified their approach by assuming that the number of individ- uals with the advantageous allele follows the logistic differential equation. We show that the impact of a selective sweep can be accurately approximated by a random partition created by a stick-breaking process. Our simulation results show that ignoring the randomness when the number of individuals with the advantageous allele is small can lead to substantial errors. Key words: selective sweep, hitchhiking, coalescent, random partition, paintbox construction 2 When a selectively favorable mutation occurs in a population and is subsequently fixed (i.e., its frequency rises to 100%), the frequencies of alleles at closely linked loci are altered. Alleles present on the chromosome on which the original mutation occurred will tend to increase in frequency, and other alleles will decrease in frequency. Maynard Smith and Haigh (1974) referred to this as the ‘hitchhiking effect,’ because an allele can get a lift in frequency from selection acting on a neighboring allele. They considered a situation with a neutral locus with alleles A and a and a second locus where allele B has a fitness of 1 + s relative to b. -

Purebred Dog Breeds Into the Twenty-First Century: Achieving Genetic Health for Our Dogs
Purebred Dog Breeds into the Twenty-First Century: Achieving Genetic Health for Our Dogs BY JEFFREY BRAGG WHAT IS A CANINE BREED? What is a breed? To put the question more precisely, what are the necessary conditions that enable us to say with conviction, "this group of animals constitutes a distinct breed?" In the cynological world, three separate approaches combine to constitute canine breeds. Dogs are distinguished first by ancestry , all of the individuals descending from a particular founder group (and only from that group) being designated as a breed. Next they are distinguished by purpose or utility, some breeds existing for the purpose of hunting particular kinds of game,others for the performance of particular tasks in cooperation with their human masters, while yet others owe their existence simply to humankind's desire for animal companionship. Finally dogs are distinguished by typology , breed standards (whether written or unwritten) being used to describe and to recognize dogs of specific size, physical build, general appearance, shape of head, style of ears and tail, etc., which are said to be of the same breed owing to their similarity in the foregoing respects. The preceding statements are both obvious and known to all breeders and fanciers of the canine species. Nevertheless a correct and full understanding of these simple truisms is vital to the proper functioning of the entire canine fancy and to the health and well being of the animals which are the object of that fancy. It is my purpose in this brief to elucidate the interrelationship of the above three approaches, to demonstrate how distortions and misunderstandings of that interrelationship now threaten the health of all of our dogs and the very existence of the various canine breeds, and to propose reforms which will restore both balanced breed identity and genetic health to CKC breeds. -

A New Approach for Using Genome Scans to Detect Recent Positive Selection in the Human Genome
PLoS BIOLOGY A New Approach for Using Genome Scans to Detect Recent Positive Selection in the Human Genome Kun Tang1*, Kevin R. Thornton2, Mark Stoneking1 1 Department of Evolutionary Genetics, Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany, 2 Department of Ecology and Evolutionary Biology, University of California Irvine, Irvine, California, United States of America Genome-wide scanning for signals of recent positive selection is essential for a comprehensive and systematic understanding of human adaptation. Here, we present a genomic survey of recent local selective sweeps, especially aimed at those nearly or recently completed. A novel approach was developed for such signals, based on contrasting the extended haplotype homozygosity (EHH) profiles between populations. We applied this method to the genome single nucleotide polymorphism (SNP) data of both the International HapMap Project and Perlegen Sciences, and detected widespread signals of recent local selection across the genome, consisting of both complete and partial sweeps. A challenging problem of genomic scans of recent positive selection is to clearly distinguish selection from neutral effects, given the high sensitivity of the test statistics to departures from neutral demographic assumptions and the lack of a single, accurate neutral model of human history. We therefore developed a new procedure that is robust across a wide range of demographic and ascertainment models, one that indicates that certain portions of the genome clearly depart from neutrality. Simulations of positive selection showed that our tests have high power towards strong selection sweeps that have undergone fixation. Gene ontology analysis of the candidate regions revealed several new functional groups that might help explain some important interpopulation differences in phenotypic traits. -

Refining the Use of Linkage Disequilibrium As A
HIGHLIGHTED ARTICLE | INVESTIGATION Refining the Use of Linkage Disequilibrium as a Robust Signature of Selective Sweeps Guy S. Jacobs,*,†,1 Timothy J. Sluckin,* and Toomas Kivisild‡ *Mathematical Sciences, University of Southampton, Southampton SO17 1BJ, United Kingdom, †Complexity Institute, Nanyang Technological University, Singapore 637723, and ‡Department of Biological Anthropology, University of Cambridge, Cambridge CB2 1QH, United Kingdom ORCID IDs: 0000-0002-4698-7758 (G.S.J.); 0000-0002-9163-0061 (T.J.S.); 0000-0002-6297-7808 (T.K.) ABSTRACT During a selective sweep, characteristic patterns of linkage disequilibrium can arise in the genomic region surrounding a selected locus. These have been used to infer past selective sweeps. However, the recombination rate is known to vary substantially along the genome for many species. We here investigate the effectiveness of current (Kelly’s ZnS and vmax) and novel statistics at inferring hard selective sweeps based on linkage disequilibrium distortions under different conditions, including a human-realistic demographic model and recombination rate variation. When the recombination rate is constant, Kelly’s ZnS offers high power, but is outperformed by a novel statistic that we test, which we call Za: We also find this statistic to be effective at detecting sweeps from standing variation. When recombination rate fluctuations are included, there is a considerable reduction in power for all linkage disequilibrium-based statistics. However, this can largely be reversed by appropriately controlling for expected linkage disequilibrium using a genetic map. To further test these different methods, we perform selection scans on well-characterized HapMap data, finding that all three statistics—vmax; Kelly’s ZnS; and Za—are able to replicate signals at regions previously identified as selection candidates based on population differentiation or the site frequency spectrum. -

Blekinge Kennelklubbs Nordic Dog Show 8-9 Augusti 2020
BLEKINGE KENNELKLUBBS NORDIC DOG SHOW 8-9 AUGUSTI 2020. Utställningsplats/Showground: Ronneby Brunnspark Sista anmälningsdag för anmälan per post utgår 10 juli/Last entry by post July 10 Sista anmälningsdag via internetanmälan utgår 14 juli, kl 12.00 /Last entry via SKK online entry July 14, 12.00 pm (noon). Anmäl via SKKs Internetanmälan/Enter via SKKs entry online: www.skk.se. In order to use the online entry the dog has to be in SKKs data base. For foreign dogs please send a copy of the registration certificate/pedigree to [email protected] a few days before you want to make the entry online. Internetanmälan gäller endast utställning, ej juniorhandling/Online entry for shows only not juniorhandling. Anmälan per post/Enter by post: blanketten/entry form ”Tävlingsanmälan” (finns att hämta på/download at www.skk.se) skickas till/send to Blekinge Kennelklubb, Västraprinsgatan 27 A, 371 35 Karlskrona, tel +46(0)709 625 662. Avgiften sätts in på Blekinge Kennelklubbs BankGiro 588-8649. Frågor rörande din anmälan, kontakta/Inquiries regarding your entry: Jonas Johansson, telefon +46(0)709 625 662, e-post [email protected]. Allmänna upplysningar/information: Tel. +46 733 24 51 41, e-post [email protected]. Du hittar oss även på/You may also look at www.skk.se/bkk/ eller kontakta Svenska Kennelklubben, Tävlingsavdelningen, 08-795 33 22, 10-12, 13-15. Payment: Please pay the entry fee via SWIFT to our IBAN account number SE 3650000000056811019662, Skandinaviska enskilda banken, Karlskrona, Sweden. SWIFT address: ESSESESS Payment receipts must indicate the name of the dog, registration number and owner. -

Identification of Selective Sweeps, Major Genes, and Genotype by Diet Interactions Melanie D
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Theses and Dissertations in Animal Science Animal Science Department 12-2015 Genomic Analysis of Sow Reproductive Traits: Identification of Selective Sweeps, Major Genes, and Genotype by Diet Interactions Melanie D. Trenhaile University of Nebraska-Lincoln, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/animalscidiss Part of the Meat Science Commons Trenhaile, Melanie D., "Genomic Analysis of Sow Reproductive Traits: Identification of Selective Sweeps, Major Genes, and Genotype by Diet Interactions" (2015). Theses and Dissertations in Animal Science. 114. http://digitalcommons.unl.edu/animalscidiss/114 This Article is brought to you for free and open access by the Animal Science Department at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Theses and Dissertations in Animal Science by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. GENOMIC ANALYSIS OF SOW REPRODUCTIVE TRAITS: IDENTIFICATION OF SELECTIVE SWEEPS, MAJOR GENES, AND GENOTYPE BY DIET INTERACTIONS By Melanie Dawn Trenhaile A THESIS Presented to the Faculty of The Graduate College at the University of Nebraska In Partial Fulfillment of Requirements For the Degree of Master of Science Major: Animal Science Under the Supervision of Professor Daniel Ciobanu Lincoln, Nebraska December, 2015 GENOMIC ANALYSIS OF SOW REPRODUCTIVE TRAITS: IDENTIFICATION OF SELECTIVE SWEEPS, MAJOR GENES, AND GENOTYPE BY DIET INTERACTIONS Melanie D. Trenhaile, M.S. University of Nebraska, 2015 Advisor: Daniel Ciobanu Reproductive traits, such as litter size and reproductive longevity, are economically important. However, selection for these traits is difficult due to low heritability, polygenic nature, sex-limited expression, and expression late in life. -
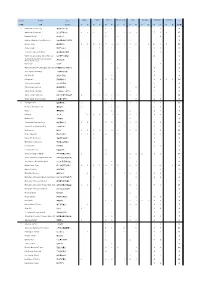
Group Breed Class Baby Puppy Junior Intermediate Open Working
Baby Puppy Junior Intermediate Open Working Champion Veteran Group Breed Class ∑ Gender D B D B D B D B D B D B D B D B 犬种组 犬种 总只数 1 Australian Cattle Dog 2 1 1 4 澳大利亚牧牛犬 Australian Shepherd 1 1 1 1 1 1 1 7 2 1 17 澳大利亚牧羊犬 Bearded Collie 1 1 2 4 长须牧羊犬 Belgian Shepherd Dog (Malinois) 1 1 玛丽诺斯比利时牧羊犬 Border Collie 2 3 2 1 7 4 2 1 7 5 1 10 8 2 55 边境牧羊犬 Collie Rough 1 1 1 1 1 5 苏格兰牧羊犬 Czechoslovakian Wolfdog 2 2 1 5 捷克斯洛伐克狼犬 Dutch Shepherd Dog (Short-Haired) ( ) 1 1 荷兰牧羊犬 ~v German Shepherd Dog (Long And 1 1 Harsh Outer Coat) 德国牧羊犬 Komondor 1 1 2 可蒙犬 Maremma And The Abruzzes Sheepdog 1 1 2 玛瑞玛安布卢斯牧羊犬 Old English Sheepdog 1 1 古代英国牧羊犬 Puli (Black) ( ) 1 1 2 波利犬 e? Schipperke 2 2 2 2 1 1 10 西帕凯牧羊犬 Shetland Sheepdog 1 2 1 5 9 喜乐蒂牧羊犬 Tatra Shepherd Dog 1 1 2 泰托拉牧羊犬 Welsh Corgi Cardigan 1 2 3 卡迪根威尔士柯基犬 Welsh Corgi Pembroke 4 1 1 12 9 2 3 1 11 5 49 彭布罗克威尔士柯基犬 White Swiss Shepherd Dog 1 3 2 2 1 9 白色瑞士牧羊犬 2 Affenpinscher 1 2 3 1 4 2 13 猴面宾莎犬 Bernese Mountain Dog 2 1 2 2 1 8 伯恩山犬 Boxer 1 2 1 5 9 德国拳师犬 Bulldog 1 2 1 4 3 2 2 1 8 1 25 斗牛犬 Bullmastiff 1 1 斗牛獒犬 Caucasian Shepherd Dog 1 2 2 2 1 1 3 2 14 高加索牧羊犬 Central Asia Shepherd Dog 1 1 1 1 2 6 中亚牧羊犬 Dobermann 1 4 2 3 9 4 2 3 2 9 4 43 杜宾犬 Dogo Argentino 1 1 1 3 1 1 3 2 13 阿根廷杜高犬 Dogue De Bordeaux 1 1 2 法国波尔多獒犬 Entlebuch Cattle Dog 1 1 恩特雷布赫牧牛犬 Fila Brasileiro 1 1 2 巴西獒犬 German Pinscher 1 1 德国宾莎犬 Giant Schnauzer (Black) ( ) 1 1 2 2 1 1 2 10 巨型雪c$犬 e? Giant Schnauzer (Pepper And Salt) ( ) 1 1 巨型雪c$犬 椒盐? Great Dane (Black/Harlequin) ( / ) 1 1 2 大丹犬 e 黑白花色 Italian Cane Corso 2 1 2 2 4 1 3 2 3 6 4 1 31 意大利凯因克尔索犬 Majorca Mastiff 1 1 2 马略卡獒犬 Miniature Pinscher 1 1 1 1 1 3 1 9 迷你宾莎犬 Miniature Schnauzer (Black And Silver) ( ) 1 1 1 1 4 ]^雪c$犬 黑银? Miniature Schnauzer (Black) ( ) 2 2 1 1 1 7 ]^雪c$犬 e? Miniature Schnauzer (Pepper And Salt) ( ) 1 2 9 3 4 1 5 2 27 ]^雪c$犬 椒盐? Miniature Schnauzer (White) ( ) 1 1 2 ]^雪c$犬 >? Newfoundland 1 1 纽芬兰犬 Presa Canario 1 1 1 3 普雷萨加纳利犬 Rottweiler 1 1 2 1 1 1 1 1 9 罗威纳犬 Russian Black Terrier 1 1 2 1 2 7 俄罗斯黑更犬 Shar Pei 1 2 1 4 沙皮犬 St. -

Dog Breeds Pack 1 Professional Vector Graphics Page 1
DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 1 Affenpinscher Afghan Hound Aidi Airedale Terrier Akbash Akita Inu Alano Español Alaskan Klee Kai Alaskan Malamute Alpine Dachsbracke American American American American Akita American Bulldog Cocker Spaniel Eskimo Dog Foxhound American American Mastiff American Pit American American Hairless Terrier Bull Terrier Staffordshire Terrier Water Spaniel Anatolian Anglo-Français Appenzeller Shepherd Dog de Petite Vénerie Sennenhund Ariege Pointer Ariegeois COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 2 Armant Armenian Artois Hound Australian Australian Kelpie Gampr dog Cattle Dog Australian Australian Australian Stumpy Australian Terrier Austrian Black Shepherd Silky Terrier Tail Cattle Dog and Tan Hound Austrian Pinscher Azawakh Bakharwal Dog Barbet Basenji Basque Basset Artésien Basset Bleu Basset Fauve Basset Griffon Shepherd Dog Normand de Gascogne de Bretagne Vendeen, Petit Basset Griffon Bavarian Mountain Vendéen, Grand Basset Hound Hound Beagle Beagle-Harrier COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 2 PROFESSIONAL VECTOR GRAPHICS PAGE 3 Belgian Shepherd Belgian Shepherd Bearded Collie Beauceron Bedlington Terrier (Tervuren) Dog (Groenendael) Belgian Shepherd Belgian Shepherd Bergamasco Dog (Laekenois) Dog (Malinois) Shepherd Berger Blanc Suisse Berger Picard Bernese Mountain Black and Berner Laufhund Dog Bichon Frisé Billy Tan Coonhound Black and Tan Black Norwegian -

Crossbreeding Systems for Small Beef Herds
~DMSION OF AGRICULTURE U~l_}J RESEARCH &: EXTENSION Agriculture and Natural Resources University of Arkansas System FSA3055 Crossbreeding Systems for Small Beef Herds Bryan Kutz For most livestock species, Hybrid Vigor Instructor/Youth crossbreeding is an important aspect of production. Intelligent crossbreed- Generating hybrid vigor is one of Extension Specialist - the most important, if not the most ing generates hybrid vigor and breed Animal Science important, reasons for crossbreeding. complementarity, which are very important to production efficiency. Any worthwhile crossbreeding sys- Cattle breeders can obtain hybrid tem should provide adequate levels vigor and complementarity simply by of hybrid vigor. The highest level of crossing appropriate breeds. However, hybrid vigor is obtained from F1s, sustaining acceptable levels of hybrid the first cross of unrelated popula- vigor and breed complementarity in tions. To sustain F1 vigor in a herd, a a manageable way over the long term producer must avoid backcrossing – requires a well-planned crossbreed- not always an easy or a practical thing ing system. Given this, finding a way to do. Most crossbreeding systems do to evaluate different crossbreeding not achieve 100 percent hybrid vigor, systems is important. The following is but they do maintain acceptable levels a list of seven useful criteria for evalu- of hybrid vigor by limiting backcross- ating different crossbreeding systems: ing in a way that is manageable and economical. Table 1 (inside) lists 1. Merit of component breeds expected levels of hybrid vigor or het- erosis for several crossbreeding sys- 2. Hybrid vigor tems. 3. Breed complementarity 4. Consistency of performance Definitions 5. Replacement considerations hybrid vigor – an increase in 6. -

Judge Instruction 2021
23 December 2020 JN / Engdom21.doc JUDGE INSTRUCTION 2021 for International and Nordic dog shows organised by the DKK Please read this instruction very carefully, as changes or new rules may have been introduced since the last time you judged in Denmark. General information: A judge may not enter a dog registered in his/her name at a show where he/she is officiating as a judge. A judge may not handle any dog at a show where he/she is officiating as a judge. The above rules apply to all competitions at the show in question. Exempted from the above are judges who judge junior handling competitions only. These judges are permitted to enter/show/handle dogs at the show in question, but they are not permitted to take part in the competitions in the Main Ring. A partner, any member of his/her immediate family or any person living with him/her in his/her household may enter and handle any dog (provided that the dog is not registered in the judge’s name) of such breed(s), which this judge is not judging on that day of the show. The dogs that the judge handles at a Dansk Kennel Klub dog show, where he/she is not acting as a judge must be either bred, owned or co-owned by him/her, a partner, a member of his/her imme- diate family or any person living with him/her in his/her household, or the judge must have dis- posal of the dog. In the last-mentioned case, a copy of the declaration of disposal must be en- closed when the dog is entered for the show. -

11. Oktober 2020 Slovenia, Celje SPLOŠNE INFORMACIJE / GENERAL INFORMATION / ALLGEMEINE INFORMATIONEN
8.-11. oktober 2020 Slovenia, Celje SPLOŠNE INFORMACIJE / GENERAL INFORMATION / ALLGEMEINE INFORMATIONEN Eurodogshow 2020 Kinološka zveza Slovenije Organizator/Organiser Zapoge 3d SI-1217 Vodice www.eurodogshow2020.eu Kontakt/Contact [email protected] facebook: Euro Dogshow 2020 - Feel Slovenia Predsednik organizacijskega odbora [email protected] President of the organising comitee Strokovni vodja razstave [email protected] Expert Manager Marketing [email protected] Novinarji/Press [email protected] RAZPORED PO DNEVIH/ DISTRIBUTION BY DAY/EINTEILUNG DER FCI GRUPPEN četrtek/Thursday/ petek/Friday/ sobota/Saturday/ nedelja/Sunday/ Donnerstag Freitag Samstag Sonntag 8.10.2020 9.10.2020 10.10.2020 11.10.2020 FCI III FCI I FCI II CACIB FCI VII FCI V FCI IV SLOVENIAN WINNER SHOW FCI VIII FCI IX FCI VI FCI X DRUGE INFORMACIJE/ OTHER INFORMATION/ANDERE INFORMATIONEN Celjski Sejem Lokacija/Location of show Dečkova ulica 1 SI-3000 Celje Prijave bodo odprte konec decembra 2019. Prijave/Entries Entries will be open end of December 2019. Za vse pasme bodo organizirane specialne razstave, za pasme ki nimajo klubov ali le ti ne bodo organizirali specialnih razstav, bo le-te organizirala KZS. Informacije bodo podane v januarju 2020. Specialne in Klubske razstave Specialties and Clubshows For all breeds clubshows/specialty shows will be organised. For breed withouth clubs or if clubs do not choose to organise the specialty, it will be organised by the CAS. All information will be available in January 2020.. BIS CACIB -

Rapid Evolution of a Skin-Lightening Allele in Southern African Khoesan
Rapid evolution of a skin-lightening allele in southern African KhoeSan Meng Lina,1,2, Rebecca L. Siforda,b,3, Alicia R. Martinc,d,e,3, Shigeki Nakagomef, Marlo Möllerg, Eileen G. Hoalg, Carlos D. Bustamanteh, Christopher R. Gignouxh,i,j, and Brenna M. Henna,2,4 aDepartment of Ecology and Evolution, State University of New York at Stony Brook, Stony Brook, NY 11794; bThe School of Human Evolution and Social Change, Arizona State University, Tempe, AZ 85287; cAnalytic and Translational Genetics Unit, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114; dProgram in Medical and Population Genetics, Broad Institute, Cambridge, MA 02141; eStanley Center for Psychiatric Research, Broad Institute, Cambridge, MA 02141; fSchool of Medicine, Trinity College Dublin, Dublin 2, Ireland; gDST-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town 8000, South Africa; hDepartment of Genetics, Stanford University, Stanford, CA 94305; iColorado Center for Personalized Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO 80045; and jDepartment of Biostatistics and Informatics, University of Colorado Anschutz Medical Campus, Aurora, CO 80045 Edited by Nina G. Jablonski, The Pennsylvania State University, University Park, PA, and accepted by Editorial Board Member C. O. Lovejoy October 18, 2018 (received for review February 2, 2018) Skin pigmentation is under strong directional selection in northern Amhara or another group who themselves are substantially European and Asian populations. The indigenous KhoeSan popula- admixed with Near Eastern people has been proposed (13, 14).