Evaluation of Selected Indigenous Coconut Accessions for High Nut
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Coal Directory of India 2015-16
COAL DIRECTORY OF INDIA 2015-16 Coal Statistics GOVERNMENT OF INDIA MINISTRY OF COAL COAL CONTROLLER'S ORGANISATION KOLKATA COAL DIRECTORY OF INDIA 2015-16 For any enquiry and suggestion please write to:- Coal Controller's Organisation 1, Council House Street Kolkata – 700 001 Tel: 91-33-2248-9616, 91-33-2248-8225 Fax : 91-33-2248-2459 E-mail : [email protected] Team Associated with the Publication Sri Anjani Kumar Coal Controller Sri Sudip Kr. Ray Deputy Director General Sri Anupam Lahiri Director Sri Indradeep Roy Chowdhury Deputy Director Dr. Sujoy Majumder Officer on Special Duty Sri Kingshuk Mukhopadhyay Officer on Special Duty(Project) Sri Goutam Bandyopadhyay Accountant Sri Sukumar Das Assistant Sri Sumanta Biswas UDC Miss Manashi Goswami Jr. Computor Sri Sachin Kumar LDC Mrs Ruma Nayak UDC Miss Shreya Banerjee UDC Mrs Sima Guha Ray UDC ************** Section Subject Page No. Section - 1: Historical Perspective 1.1-1.18 Overall Coal Scenario: A Review 1.1-1.9 Highlights 1.10-1.13 Chart 1.1 Trends of Productin of Primary Conventional Energy Forms in India 1.14 Table 1.1 Indian Economy - Selected Indicators 1.15 Table 1.2 Growth of Indian Coal Sector at a Glance 1.16 Table 1.3 Production of Primary Sources of Conventional Energy in India 1.17 Table 1.4 Total Praimary Supply (TPS) of Coal & Lignite 1.18 Section - 2: Resources & Exploration 2.1-2.31 Summary 2.1-2.2 Chart 2.1 Geological Coal Reserve in Major Indian Coalfields as on 01/04/2016 2.3 Chart 2.2 Gradewise Geological Reserve of Non-coking coal in Gondwana Coalfields as on -
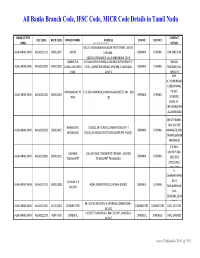
Banks Branch Code, IFSC Code, MICR Code Details in Tamil Nadu
All Banks Branch Code, IFSC Code, MICR Code Details in Tamil Nadu NAME OF THE CONTACT IFSC CODE MICR CODE BRANCH NAME ADDRESS CENTRE DISTRICT BANK www.Padasalai.Net DETAILS NO.19, PADMANABHA NAGAR FIRST STREET, ADYAR, ALLAHABAD BANK ALLA0211103 600010007 ADYAR CHENNAI - CHENNAI CHENNAI 044 24917036 600020,[email protected] AMBATTUR VIJAYALAKSHMIPURAM, 4A MURUGAPPA READY ST. BALRAJ, ALLAHABAD BANK ALLA0211909 600010012 VIJAYALAKSHMIPU EXTN., AMBATTUR VENKATAPURAM, TAMILNADU CHENNAI CHENNAI SHANKAR,044- RAM 600053 28546272 SHRI. N.CHANDRAMO ULEESWARAN, ANNANAGAR,CHE E-4, 3RD MAIN ROAD,ANNANAGAR (WEST),PIN - 600 PH NO : ALLAHABAD BANK ALLA0211042 600010004 CHENNAI CHENNAI NNAI 102 26263882, EMAIL ID : CHEANNA@CHE .ALLAHABADBA NK.CO.IN MR.ATHIRAMIL AKU K (CHIEF BANGALORE 1540/22,39 E-CROSS,22 MAIN ROAD,4TH T ALLAHABAD BANK ALLA0211819 560010005 CHENNAI CHENNAI MANAGER), MR. JAYANAGAR BLOCK,JAYANAGAR DIST-BANGLAORE,PIN- 560041 SWAINE(SENIOR MANAGER) C N RAVI, CHENNAI 144 GA ROAD,TONDIARPET CHENNAI - 600 081 MURTHY,044- ALLAHABAD BANK ALLA0211881 600010011 CHENNAI CHENNAI TONDIARPET TONDIARPET TAMILNADU 28522093 /28513081 / 28411083 S. SWAMINATHAN CHENNAI V P ,DR. K. ALLAHABAD BANK ALLA0211291 600010008 40/41,MOUNT ROAD,CHENNAI-600002 CHENNAI CHENNAI COLONY TAMINARASAN, 044- 28585641,2854 9262 98, MECRICAR ROAD, R.S.PURAM, COIMBATORE - ALLAHABAD BANK ALLA0210384 641010002 COIIMBATORE COIMBATORE COIMBOTORE 0422 2472333 641002 H1/H2 57 MAIN ROAD, RM COLONY , DINDIGUL- ALLAHABAD BANK ALLA0212319 NON MICR DINDIGUL DINDIGUL DINDIGUL -

Coal & Lignite
COAL & LIGNITE Indian Minerals Yearbook 2014 (Part- III : Mineral Reviews) 53rd Edition COAL & LIGNITE (FINAL RELEASE) GOVERNMENT OF INDIA MINISTRY OF MINES INDIAN BUREAU OF MINES Indira Bhavan, Civil Lines, NAGPUR – 440 001 PHONE/FAX NO. (0712) 2565471 PBX : (0712) 2562649, 2560544, 2560648 E-MAIL : [email protected] Website: www.ibm.gov.in July, 2016 10-1 COAL & LIGNITE 10 Coal & Lignite oal plays a pivotal role in sustainable development. Meghalaya. As a result of exploration carried out by CIt is the most widely used energy source for GSI, CMPDIL and other agencies, 301.56 billion electricity generation and an essential input for steel tonnes (including that estimated in Sikkim) coal production. Coal is an essential resource for meeting reserves up to 1,200 m depth have been established the challenges facing the modern world. As per in the country as on 1.4.2014. Out of these Integrated Energy Policy Committee of Planning reserves, 125.91 billion tonnes are proved reserves, Commission, coal will remain India's most important 142.50 billion tonnes are indicated reserves and the energy source till 2031-32 and possibly beyond. In remaining 33.15 billion tonnes are in inferred India, about 76% coal output is consumed in power category. Of the total reserves, the share of sector. In addition, other industries like cement, prime-coking coal is 5.31 billion tonnes, medium- fertilizer, chemical, paper and thousands of medium and coking & semi-coking is 28.76 billion tonnes and small-scale industries are dependent on coal for their non-coking coal, including high sulphur is process and energy requirements. -
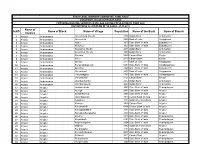
S.NO Name of District Name of Block Name of Village Population Name
STATE LEVEL BANKERS' COMMITTEE, TAMIL NADU CONVENOR: INDIAN OVERSEAS BANK PROVIDING BANKING SERVICES IN VILLAGE HAVING POPULATION OF OVER 2000 DISTRICTWISE ALLOCATION OF VILLAGES -01.11.2011 Name of S.NO Name of Block Name of Village Population Name of the Bank Name of Branch District 1 Ariyalur Andiamadam Anikudichan (South) 2730 Indian Bank Andimadam 2 Ariyalur Andiamadam Athukurichi 5540 Bank of India Alagapuram 3 Ariyalur Andiamadam Ayyur 3619 State Bank of India Edayakurichi 4 Ariyalur Andiamadam Kodukkur 3023 State Bank of India Edayakurichi 5 Ariyalur Andiamadam Koovathur (North) 2491 Indian Bank Andimadam 6 Ariyalur Andiamadam Koovathur (South) 3909 Indian Bank Andimadam 7 Ariyalur Andiamadam Marudur 5520 Canara Bank Elaiyur 8 Ariyalur Andiamadam Melur 2318 Canara Bank Elaiyur 9 Ariyalur Andiamadam Olaiyur 2717 Bank of India Alagapuram 10 Ariyalur Andiamadam Periakrishnapuram 5053 State Bank of India Varadarajanpet 11 Ariyalur Andiamadam Silumbur 2660 State Bank of India Edayakurichi 12 Ariyalur Andiamadam Siluvaicheri 2277 Bank of India Alagapuram 13 Ariyalur Andiamadam Thirukalappur 4785 State Bank of India Varadarajanpet 14 Ariyalur Andiamadam Variyankaval 4125 Canara Bank Elaiyur 15 Ariyalur Andiamadam Vilandai (North) 2012 Indian Bank Andimadam 16 Ariyalur Andiamadam Vilandai (South) 9663 Indian Bank Andimadam 17 Ariyalur Ariyalur Andipattakadu 3083 State Bank of India Reddipalayam 18 Ariyalur Ariyalur Arungal 2868 State Bank of India Ariyalur 19 Ariyalur Ariyalur Edayathankudi 2008 State Bank of India Ariyalur 20 Ariyalur -

Trade Marks Journal No: 1841, 19/03/2018 P`Kasana : Baart Sarkar
Trade Marks Journal No: 1841, 19/03/2018 Reg. No. TECH/47-714/MBI/2000 Registered as News Paper p`kaSana : Baart sarkar vyaapar icanh rijasT/I esa.ema.raoD eMTa^p ihla ko pasa paosT Aa^ifsa ko pasa vaDalaa mauMba[- 400037 durBaaYa : 022 24101144 ,24101177 ,24148251 ,24112211. fO@sa : 022 24140808 Published by: The Government of India, Office of The Trade Marks Registry, Baudhik Sampada Bhavan (I.P. Bhavan) Near Antop Hill, Head Post Office, S.M. Road, Mumbai-400037. Tel:022-24140808 1 Trade Marks Journal No: 1841, 19/03/2018 Anauk/maiNaka INDEX AiQakairk saucanaaeM Official Notes vyaapar icanh rijasT/IkrNa kayaa-laya ka AiQakar xao~ Jurisdiction of Offices of the Trade Marks Registry sauiBannata ko baaro maoM rijaYT/ar kao p`arMiBak salaah AaoOr Kaoja ko ilayao inavaodna Preliminary advice by Registrar as to distinctiveness and request for search saMbaw icanh Associated Marks ivaraoQa Opposition ivaiQak p`maaNa p`~ iT.ema.46 pr AnauraoQa Legal Certificate/ Request on Form TM-46 k^apIra[T p`maaNa p`~ Copyright Certificate t%kala kaya- Operation Tatkal saava-jainak saucanaaeM Public Notices svaIkRit ko puva- iva&aipt Aavaodna Applications advertised before acceptance-class-wise: 2 Trade Marks Journal No: 1841, 19/03/2018 vaga- / Class - 1 11-165 vaga- / Class - 2 166-208 vaga- / Class - 3 209-587 vaga- / Class - 4 588-624 vaga- / Class - 5 625-2037 vaga- / Class - 6 2038-2131 vaga- / Class - 7 2132-2344 vaga- / Class - 8 2345-2378 vaga- / Class - 9 2379-2867 vaga- / Class - 10 2868-2957 vaga- / Class - 11 2958-3154 vaga- / Class - 12 3155-3252 -

Coal & Lignite
COAL & LIGNITE Indian Minerals Yearbook 2019 (Part- III : MINERAL REVIEWS) 58th Edition COAL & LIGNITE (ADVANCE RELEASE) GOVERNMENT OF INDIA MINISTRY OF MINES INDIAN BUREAU OF MINES Indira Bhavan, Civil Lines, NAGPUR – 440 001 PHONE/FAX NO. (0712) 2565471 PBX: (0712) 2562649, 2560544, 2560648 E-MAIL: [email protected] Website: www.ibm.gov.in August, 2020 7- 1 COAL & LIGNITE 7 Coal & Lignite oal is a fossil fuel. It is a combustible, 728.72 million tonnes in 2018-19. The production of C sedimentary, organic rock, which is composed lignite at 44.28 million tonnes in 2018-19 decreased by mainly of carbon, hydrogen and oxygen. It is formed about 5.1% from 46.64 million tonnes in the previous from vegetation, which has been consolidated between year. India, in 2018 ranked 2nd in the world coal rock strata and altered by the combined effects of production. pressure and heat over millions of years to form coal seams. GEOLOGICAL RESOURCES The build-up of silt and other sediments, together Coal with movements in the earth’s crust (known as tectonic The coal deposits in India primarily are movements) buried these swamps and peat bogs, often concentrated in the Gondwana sediments occurring to great depths causing the plant material to be mainly in the eastern and central parts of Peninsular subjected to high temperatures and pressures. Millions India, although Gondwana coal deposits also are found of year of deep burial engendered such physical and to occur in the north-eastern part of the country mainly chemical changes which transformed the vegetation in Assam and Sikkim. -

Coal Directory of India 2017-18
COAL DIRECTORY OF INDIA 2017-18 [ Coal Statistics ] GOVERNMENT OF INDIA MINISTRY OF COAL COAL CONTROLLER'S ORGANISATION KOLKATA COAL DIRECTORY OF INDIA 2017-18 For any enquiry and suggestion please write to:- Coal Controller's Organisation 1, Council House Street Kolkata – 700 001 Tel: 91-33-2248-9616, 91-33-2248-8225 Fax : 91-33-2248-2459 E-mail : [email protected] efasdfs Team Associated with the Publication Sri Anjani Kumar Coal Controller Sri Vishwanath Pratap Singh Joint Director, I.S.S. Sri Indradeep Roy Chowdhury Deputy Director, I.S.S. Dr. Sujoy Majumder Dy. G. M. (SCCL)/OSD Sri Kingshuk Mukhopadhyay Sr. Manager (ECL)/OSD Sri Sukumar Das Superintendent Sri Sumanta Biswas Assistant Sri Chandan Bandopadhyay Assistant Mrs Sima Guha Ray Assistant Mrs Ruma Nayak UDC Miss Manashi Goswami Computor Operator Sri Amit Kumar LDC ************** Section Subject Page No. Section - 1: Historical Perspective 1.1-1.17 Overall Coal Scenario: A Review 1.1-1.9 Highlights 1.10-1.13 Chart 1.1 Trends of Production of Primary Conventional Energy Forms in India 1.14 Table 1.1 Growth of Indian Coal Sector at a Glance 1.15 Table 1.2 Production of Primary Sources of Conventional Energy in India 1.16 Table 1.3 Total Praimary Supply (TPS) of Coal & Lignite 1.17 Section - 2: Resources & Exploration 2.1-2.31 Summary 2.1-2.2 Chart 2.1 Geological Coal Reserve in Major Indian Coalfields as on 01/04/2018 2.3 Chart 2.2 Gradewise Geological Reserve of Non-coking coal in Gondwana Coalfields as on 01/04/2018 2.3 Chart 2.3 Statewise Geological Reserve of Indian Coal -

Thanjavur District
CENSUS OF INDIA 2011 TOTAL POPULATION AND POPULATION OF SCHEDULED CASTES AND SCHEDULED TRIBES FOR VILLAGE PANCHAYATS AND PANCHAYAT UNIONS THANJAVUR DISTRICT DIRECTORATE OF CENSUS OPERATIONS TAMILNADU ABSTRACT THANJAVUR DISTRICT No. of Total Total Sl. No. Panchayat Union Total Male Total SC SC Male SC Female Total ST ST Male ST Female Village Population Female 1 Thanjavur 61 2,30,116 1,14,931 1,15,185 52,012 25,935 26,077 230 115 115 2 Budalur 42 91,552 45,519 46,033 26,874 13,396 13,478 43 19 24 3 Thiruvaiyaru 40 98,089 48,795 49,294 27,201 13,513 13,688 19 10 9 4 Orathanadu 58 1,60,367 77,719 82,648 23,127 11,242 11,885 55 30 25 5 Thiruvonam 30 86,953 42,685 44,268 20,434 10,132 10,302 20 9 11 6 Kumbakonam 47 1,84,611 91,944 92,667 50,478 25,405 25,073 214 107 107 7 Thiruvidaimarudur 48 1,33,215 66,400 66,815 30,794 15,416 15,378 107 51 56 8 Thiruppanandal 44 1,04,663 52,286 52,377 42,267 21,216 21,051 278 137 141 9 Papanasam 34 1,16,975 56,622 60,353 27,368 13,577 13,791 80 36 44 10 Ammapettai 46 1,00,022 49,911 50,111 37,148 18,534 18,614 99 51 48 11 Pattukkottai 43 1,13,231 54,227 59,004 16,708 8,017 8,691 94 45 49 12 Madukkur 33 67,113 31,414 35,699 8,789 4,196 4,593 25 17 8 13 Peravurani 26 89,164 43,813 45,351 11,796 5,769 6,027 179 93 86 14 Sethubhavachatram 37 91,738 44,617 47,121 9,295 4,632 4,663 22 11 11 Grand Total 589 16,67,809 8,20,883 8,46,926 3,84,291 1,90,980 1,93,311 1,465 731 734 THANJAVUR PANCHAYAT UNION Sl. -

Tamil Nadu Government Gazette
© [Regd. No. TN/CCN/467/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 197/2009. 2015 [Price: Rs. 35.20 Paise. TAMIL NADU GOVERNMENT GAZETTE PUBLISHED BY AUTHORITY No. 2] CHENNAI, WEDNESDAY, JANUARY 14, 2015 Margazhi 30, Jaya, Thiruvalluvar Aandu – 2045 Part VI—Section 4 Advertisements by private individuals and private institutions CONTENTS PRIVATE ADVERTISEMENTS Pages. Change of Names .. 61-147 Notice .. 147-148 NOTICE NO LEGAL RESPONSIBILITY IS ACCEPTED FOR THE PUBLICATION OF ADVERTISEMENTS REGARDING CHANGE OF NAME IN THE TAMIL NADU GOVERNMENT GAZETTE. PERSONS NOTIFYING THE CHANGES WILL REMAIN SOLELY RESPONSIBLE FOR THE LEGAL CONSEQUENCES AND ALSO FOR ANY OTHER MISREPRESENTATION, ETC. (By Order) Director of Stationery and Printing. CHANGE OF NAMES 878. I, J Jeyakanthan, son of Thiru D Jagadeeswaran, 881. I, K. Muniyandi, son of Thiru M. Kamalamuthu, born on born on 24th June 1967 (native district: Tiruvallur), residing 4th December 1969 (native district: Madurai), residing at at No. HIG-3, May Flower Garden, Srinivasa Avenue Road, No. 10, Mahal 3rd Lane, M.K. Lane, Manjanakara Street, Raja Annamalaipuram, Chennai-600 028, shall henceforth South Masi Vethi, Madurai-625 001, shall henceforth be be known as J JAYAKANTHAN. known as ARAVINTH KARUTAKAR. ªü ªüò裉î¡. K. MUNIYANDI. Chennai, 12th January 2015. Madurai, 5th January 2015. 879. I, N. Mohan, son of Thiru P. Nambirajan, born on 882. I, K. Muthu, son of Thiru V. Kanda Nadar, born on 18th April 1996 (native district: Tirunelveli), residing at 1st January 1961 (native district: Theni), residing at Old No.15, New No. 16/1/E, Chidambarapuram, Rajapudur Old No. -

Tamil Nadu Government Gazette
© [Regd. No. TN/CCN/467/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 197/2009. 2021 [Price: Rs. 128.00 Paise. TAMIL NADU GOVERNMENT GAZETTE PUBLISHED BY AUTHORITY No. 7A] CHENNAI, WEDNESDAY, FEBRUARY 17, 2021 Maasi 5, Saarvari, Thiruvalluvar Aandu–2052 Part II—Section 2 (Supplement) NOTIFICATIONS BY GOVERNMENT AGRICULTURE DEPARTMENT Secretariat, 17th February 2021. Notifi cation of Crops under Revamped Pradhan Mantri Fasal Bima Yojana during Special Season 2020-2021 [AGRICULTURE – NOTIFICATION OF CROPS (PADDY II (SAMBA / THALADI / PISHANAM), MAIZE II, COTTON II & ONION II) AND INSURANCE UNITS FOR IMPLEMENTATION OF REVAMPED PRADHAN MANTRI FASAL BIMA YOJANA (RPMFBY) DURING SPECIAL SEASON, 2020-2021 – PUBLICATION IN THE TAMIL NADU GOVERNMENT GAZETTE – ORDERS – ISSUED.] The Following Government Order is Published:— [G.O. Ms. No. 253, Agriculture (AP2), 4th November 2020, äŠðC 19, ꣘õK, F¼õœÀõ˜ ݇´-2051.] No. II(2)/AG/108/2021. R:- 1. G.O.(Ms.) No.178, Agriculture (AP2) Department, dated 07.08.2020. 2. G.O.(D) No.141, Agriculture (AP2) Department dated 14.08.20. 3. G.O.(D).No.178, Agriculture (AP2) Department, dated 03.10.2020. 4. Government Letter No.18080/AP2/2020, Agriculture (AP2) Department, dated 13.10.2020. 5. Minutes of 47th State Level Coordination Committee on Crop Insurance (SLCCCI) meeting held on 20.10.2020 for notifi cation of Crops and Insurance Units under Revamped Pradhan Mantri Fasal Bima Yojana (RPMFBY) during Special Season 2020-2021. 6. From the Director of Agriculture Letter No.CIS1/99164/ 2020, dated 22.10.2020. 7. Government D.O.letter No.19026/AP2/2020-1, Agriculture (AP2) Department, dated 27.10.2020. -

Trade Marks Journal No: 1841 , 19/03/2018 Class 28 2107454 01
Trade Marks Journal No: 1841 , 19/03/2018 Class 28 2107454 01/03/2011 ADITYA BIRLA RETAIL LIMITED 5TH AND 6TH FLOORS,SKYLINE ICON, 86/92, NEAR MITTAL INDUSTRIAL ESTATE, ANDHERI KURLA ROAD, ANDHERI (EAST), MUMBAI-400059 MANUFACTUPER, MARCHANTS AND TRADERS A COMPANY REGISTERED UNDER THE COMPANIES ACT,1956 Proposed to be Used MUMBAI GAMES AND PLAYTHINGS, GYMNASTIC AND SPORTING ARTICLES NOT INCLUDED IN OTHER CLASSES; DECORATIONS FOR CHRISTMAS TREES 5076 Trade Marks Journal No: 1841 , 19/03/2018 Class 28 2107538 01/03/2011 ADITYA BIRLA RETAIL LIMITED 5th & 6th Floors,Skyline Icon,86/92,Near Mmittal Industrial Estate, Andhrei (E) MUMBAI 400 059 MANUFACTUPER, MARCHANTS AND TRADER. A COMPANY REGISTERED UNDER THE COMPANIES ACT,1956 Proposed to be Used MUMBAI GAMES AND PLAYTHINGS, GYMNASTIC AND SPORTING ARTICLES NOT INCLUDED IN OTHER CLASSES; DECORATIONS FOR CHRISTMAS TREES REGISTRATION OF THIS TRADE MARK SHALL GIVE NO RIGHT TO THE EXCLUSIVE USE OF THE other descriptive matters.. THIS IS CONDITION OF REGISTRATION THAT MARK SHALL BE USED WHOLE.. 5077 Trade Marks Journal No: 1841 , 19/03/2018 Class 28 2197529 30/08/2011 BERIWALA IMPEX PVT LTD 9081/1 MULTANI DHANDA PAHAR GUNJ NEW DELHI 110055 MANUFACTUIRING AND TRADING Address for service in India/Attorney address: GLOBAL JURIX S-191/C, 3RD FLOOR, MANAK COMPLEX, SCHOOL BLOCK, SHAKARPUR, DELHI-92 Proposed to be Used DELHI IMPORTED TOYS, EDUCATIONAL TOYS AND CHILDREN UTILITIES PRODUCTS (PLAYTHINGS) AS INCLUDED IN CLASS 28 5078 Trade Marks Journal No: 1841 , 19/03/2018 Class 28 2198590 02/09/2011 SANJAY VARSHNEY I-23/A, LAXMI NAGAR EXTENSION, DELHI-92 PRODUCTION, DEVELOPMENT & DISTRIBUTION OF GAMES AND PLAYTHINGS, GYMNASTIC AND SPORTING ARTICLES. -

S.NO Name of the Bank Name of District Name of Block Name Of
STATE LEVEL BANKERS' COMMITTEE, TAMIL NADU CONVENOR: INDIAN OVERSEAS BANK PROVIDING BANKING SERVICES IN VILLAGE HAVING POPULATION OF OVER 2000 BANKWISE ALLOCATION OF VILLAGES - 01.11.2011 S.NO Name of the Bank Name of District Name of Block Name of Village Population Name of Branch 1 Allahabad Bank Coimbatore Sultanpet Poorandampalayam 3046 Sultanpet 2 Allahabad Bank Coimbatore Sultanpet Vadavalli 2999 Sultanpet 3 Allahabad Bank Coimbatore Sultanpet Varapatti 6284 Sultanpet 4 Allahabad Bank Salem Magudanchanavadi Gudalur 5004 Magudanchavadi 5 Allahabad Bank Salem Magudanchanavadi Kanagagiri 3060 Magudanchavadi 6 Allahabad Bank Salem Magudanchanavadi Kandarkulamanickam 3198 Magudanchavadi 7 Allahabad Bank Salem Magudanchanavadi Kannanderi 4836 Magudanchavadi 8 Allahabad Bank Salem Magudanchanavadi Pudur 5912 Magudanchavadi STATE LEVEL BANKERS' COMMITTEE, TAMIL NADU CONVENOR: INDIAN OVERSEAS BANK PROVIDING BANKING SERVICES IN VILLAGE HAVING POPULATION OF OVER 2000 BANKWISE ALLOCATION OF VILLAGES - 01.11.2011 S.NO Name of the Bank Name of District Name of Block Name of Village Population Name of Branch 9 Andhra Bank Cuddalore Cuddalore Nattapattu 3437 Cuddalore 10 Andhra Bank Madurai VADIPATTI IRUMBADI KARUPPATTI 11 Andhra Bank Madurai VADIPATTI NACHIKULAM KARUPPATTI 12 Andhra Bank Namakkal Namagiripettai Mathuruthu 4800 Keeripatti 13 Andhra Bank Salem Attur Arasanatham 6267 Keeripatti 14 Andhra Bank Salem Attur Seeliampatti 3917 Keeripatti 15 Andhra Bank Sivaganga Ilayangudi Tiruvallur 2244 Ilayangudi 16 Andhra Bank Sivaganga Sakkottai