European Marine Observation and Data Network Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

High Level Environmental Screening Study for Offshore Wind Farm Developments – Marine Habitats and Species Project
High Level Environmental Screening Study for Offshore Wind Farm Developments – Marine Habitats and Species Project AEA Technology, Environment Contract: W/35/00632/00/00 For: The Department of Trade and Industry New & Renewable Energy Programme Report issued 30 August 2002 (Version with minor corrections 16 September 2002) Keith Hiscock, Harvey Tyler-Walters and Hugh Jones Reference: Hiscock, K., Tyler-Walters, H. & Jones, H. 2002. High Level Environmental Screening Study for Offshore Wind Farm Developments – Marine Habitats and Species Project. Report from the Marine Biological Association to The Department of Trade and Industry New & Renewable Energy Programme. (AEA Technology, Environment Contract: W/35/00632/00/00.) Correspondence: Dr. K. Hiscock, The Laboratory, Citadel Hill, Plymouth, PL1 2PB. [email protected] High level environmental screening study for offshore wind farm developments – marine habitats and species ii High level environmental screening study for offshore wind farm developments – marine habitats and species Title: High Level Environmental Screening Study for Offshore Wind Farm Developments – Marine Habitats and Species Project. Contract Report: W/35/00632/00/00. Client: Department of Trade and Industry (New & Renewable Energy Programme) Contract management: AEA Technology, Environment. Date of contract issue: 22/07/2002 Level of report issue: Final Confidentiality: Distribution at discretion of DTI before Consultation report published then no restriction. Distribution: Two copies and electronic file to DTI (Mr S. Payne, Offshore Renewables Planning). One copy to MBA library. Prepared by: Dr. K. Hiscock, Dr. H. Tyler-Walters & Hugh Jones Authorization: Project Director: Dr. Keith Hiscock Date: Signature: MBA Director: Prof. S. Hawkins Date: Signature: This report can be referred to as follows: Hiscock, K., Tyler-Walters, H. -

Full Text in Pdf Format
MARINE ECOLOGY PROGRESS SERIES Published September 17 Mar Ecol Prog Ser - I P P Accumulation of polychlorinated biphenyls by the inf aunal brittle stars Amphiura filiformis and A. chiajei: effects of eutrophication and selective feeding Jonas S. Gunnarsson*,Mattias Sköld** Department of Marine Ecology. Göteborg University, Kristineberg Marine Research Station. 450 34 Fiskebäckskil, Sweden ABSTRACT: Polychlonnated biphenyl (PCB) accumulation by the 2 brittle stars Amphiura filiformis and A. chjajei was studied in a laboratory experiment and in the field. In the laboratory study. the fate of '4C-2,2',4,4'-tetrachlorobiphenyl(TCB) was determined in benthic microcosn~s,with and without the addition of phytoplankton (Phaeodactylum tricornutum) Added phytoplankton was rapidly miner- alised and stimulated an increased dissolved organic carbon content in the water-column and bacterial production on the sediment surface. TCB uptake by the brittle stars was significantly higher in the microcosms enriched with phytoplankton. Differences in TCB concentrations were still significant after normalisation to lipid content, suggesting that selective feeding rather than equilibnum partitioning was the cause of the increased TCB burden. Treatment effects were more apparent in body (disk) tissue, than in the arm fraction of the bnttle stars, in agreement with the lipid content of the tissues. No difference in total organic carbon, total nitrogen or TCB concentrations of the sediment surface was detected. In the field, ophuroids and sediment cores were collected at a coastal urban estuary off the city of Göteborg, Sweden, and at an offshore station in the Kattegat Sea. Surn-PCBs of sediment and brittle stars were ca 3 times higher at the coastal station than at the offshore station. -

Bioluminescence in the Ophiuroid Amphiura Filiformis (O.F. Müller
Echinoderm Research 2001, Féral & David (eds.) ©2003 Swets <S Zeitlinger, Lisse, ISBN 90 5809 528 2 Bioluminescence in the ophiuroidAmphiura filiformis (O.F. Müller, 1776) is not temperature dependant 3 7 2 4 4 O. Bruggeman, S. Dupont, J. Mallefet Laboratoire de Physiologie Animale & Centre de Recherche sur la Biodiversité, Université catholique de Louvain, Louvain-la-Neuve, Belgium R. Bannister & M.C, Thorndyke Bourne Laboratory, Royal Halloway College, University o f London, UK ABSTRACT: Influence of temperature on the bioluminescence of the ophiuroidAmphiura filiformis was experimentally tested. Amputated arms from individuals acclimated at 6° or 14°C were incubated at five dif ferent temperatures ranging from 6° to 14°C for 2 hours before KC1 stimulation. Light response comparisons do not reveal any difference neither for the two temperatures of acclimation nor for the five temperatures of incubation. Therefore, the temperature does not affect the studied parameters of the light emission. KEYWORDS: Amphiura filiformis, ophiuroid, bio luminescence, temperature. 1 INTRODUCTION natural environment of this species, the aim of this work was to characterize the short-term (few hours) Amphiura filiformis (O.F. Müller, 1776) is a brittle star and long-term (a month) effect of temperature on the with a total diameter of 15 cm. It lives in the mud of bio luminescence A.of filiformis. North European coasts and in the Mediterranean Sea. It constitutes an important fish food resources since it has been estimated that it provides up to 301 metric 2 MATERIAL AND METHODS tons of biomass per year (Nilsson & Sköld 1996). The species is also bioluminescent and arms emit blueSpecimens o fAmphiura filiformis were collected in the light when individuals are mechanically stimulatedGullmar Fjord (Fiskebackskill, Sweden) by Petersen (Emson & Herring 1985; Mallefet pers. -
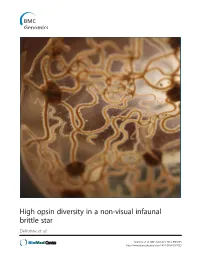
Amphiura Filiformis, We First Highlighted a Blue-Green Light Sensitivity Using a Behavioural Approach
High opsin diversity in a non-visual infaunal brittle star Delroisse et al. Delroisse et al. BMC Genomics 2014, 15:1035 http://www.biomedcentral.com/1471-2164/15/1035 Delroisse et al. BMC Genomics 2014, 15:1035 http://www.biomedcentral.com/1471-2164/15/1035 RESEARCH ARTICLE Open Access High opsin diversity in a non-visual infaunal brittle star Jérôme Delroisse1*, Esther Ullrich-Lüter2, Olga Ortega-Martinez3, Sam Dupont3, Maria-Ina Arnone4, Jérôme Mallefet5 and Patrick Flammang1 Abstract Background: In metazoans, opsins are photosensitive proteins involved in both vision and non-visual photoreception. Echinoderms have no well-defined eyes but several opsin genes were found in the purple sea urchin (Strongylocentrotus purpuratus) genome. Molecular data are lacking for other echinoderm classes although many species are known to be light sensitive. Results: In this study focused on the European brittle star Amphiura filiformis, we first highlighted a blue-green light sensitivity using a behavioural approach. We then identified 13 new putative opsin genes against eight bona fide opsin genes in the genome of S. purpuratus. Six opsins were included in the rhabdomeric opsin group (r-opsins). In addition, one putative ciliary opsin (c-opsin), showing high similarity with the c-opsin of S. purpuratus (Sp-opsin 1), one Go opsin similar to Sp-opsins 3.1 and 3.2, two basal-branch opsins similar to Sp-opsins 2 and 5, and two neuropsins similar to Sp-opsin 8, were identified. Finally, two sequences from one putative RGR opsin similar to Sp-opsin 7 were also detected. Adult arm transcriptome analysis pinpointed opsin mRNAs corresponding to one r-opsin, one neuropsin and the homologue of Sp-opsin 2. -

Echinodermata, Ophiuroidea)
Vol. 16: 105–113, 2012 AQUATIC BIOLOGY Published online July 19 doi: 10.3354/ab00435 Aquat Biol Slow arm regeneration in the Antarctic brittle star Ophiura crassa (Echinodermata, Ophiuroidea) Melody S. Clark*, Terri Souster British Antarctic Survey, Natural Environment Research Council, High Cross, Madingley Road, Cambridge CB3 0ET, UK ABSTRACT: Regeneration of arms in brittle stars is thought to proceed slowly in low temperature environments. Here a survey of natural arm damage and arm regeneration rates is documented in the Antarctic brittle star Ophiura crassa. This relatively small ophiuroid, a detritivore found amongst red macroalgae, displays high levels of natural arm damage and repair. This is largely thought to be due to ice damage in the shallow waters it inhabits. The time scale of arm regener- ation was measured in an aquarium-based 10 mo experiment. There was a delayed regeneration phase of 7 mo before arm growth was detectable in this species. This is 2 mo longer than the longest time previously described, which was in another Antarctic ophiuroid, Ophionotus victo- riae. The subsequent regeneration of arms in O. crassa occurred at a rate of approximately 0.16 mm mo−1. To date, this is the slowest regeneration rate known of any ophiuroid. The confir- mation that such a long delay before arm regeneration occurs in a second Antarctic species pro- vides strong evidence that this phenomenon is yet another characteristic feature of Southern Ocean species, along with deferred maturity, slowed growth and development rates. It is unclear whether delayed initial regeneration phases are adaptations to, or limitations of, low temperature environments. -

Fisheries Management Controls for Dab in the North Sea
Fisheries management controls for dab in The North Sea Auteurs: D.C.M. Miller and R. Verkempynck IMARES report C040/16 Fisheries management controls for dab in the North Sea Author(s): D.C.M. Miller and R. Verkempynck Client: Ministerie van Economische Zaken Directie ELVV T.a.v. Ir. H.R. Offringa Postbus 20401 2500 EK Den Haag Project nummer: BO 4311810002 BAS-code BO-20-010-076 Publication date: 12 April 2016 IMARES Wageningen UR IJmuiden, April 2016 IMARES report C040/16 D.C.M. Miller and R. Verkempynck, 2016. Fisheries management controls for dab in the North Sea; Wageningen, IMARES Wageningen UR (University & Research centre), IMARES report C040/16. © 2016 IMARES Wageningen UR IMARES, institute of Stichting DLO The Management of IMARES is not responsible for resulting damage, as well as is registered in the Dutch trade for damage resulting from the application of results or research obtained by record nr. 09098104, IMARES, its clients or any claims related to the application of information found BTW nr. NL 806511618 within its research. This report has been made on the request of the client and is wholly the client's property. This report may not be reproduced and/or published partially or in its entirety without the express written consent of the client. A_4_3_2 V21 IMARES report C040/16 | 2 van 31 Table of Contents Table of Contents ..................................................................................................................... 3 Summary ............................................................................................................................... -

1 Growth and Production of the Brittle Stars Ophiura Sarsii and Ophiocten Sericeum (Echinodermata: 2 Ophiuroidea)
1 Growth and production of the brittle stars Ophiura sarsii and Ophiocten sericeum (Echinodermata: 2 Ophiuroidea) 3 Alexandra M. Ravelo*1, Brenda Konar1, Bodil Bluhm2, Katrin Iken1 4 *(907) 474-7074 5 [email protected] 6 1School of Fisheries and Ocean Sciences, University of Alaska Fairbanks 7 P.O. Box 757220, Fairbanks, AK 99775, USA 8 2Department of Arctic and Marine Biology, University of Tromsø 9 9037 Tromsø, Norway 10 11 Abstract 12 Dense brittle star assemblages dominate vast areas of the Arctic marine shelves, making them 13 key components of Arctic ecosystem. This study is the first to determine the population dynamics of 14 the dominant shelf brittle star species, Ophiura sarsii and Ophiocten sericeum, through age determination, 15 individual production and total turnover rate (P:B). In the summer of 2013, O. sarsii were collected in 16 the northeastern Chukchi Sea (depth 35 to 65 m), while O. sericeum were collected in the central 17 Beaufort Sea (depth 37 to 200 m). Maximum age was higher for O. sarsii than for O. sericeum (27 and 18 20 years, respectively); however, both species live longer than temperate region congeners. Growth 19 curves for both species had similar initial fast growth, with an inflection period followed by a second 20 phase of fast growth. Predation avoidance in addition to changes in the allocation of energy may be 21 the mechanisms responsible for the observed age dependent growth rates. Individual production was 22 higher for O. sarsii than for O. sericeum by nearly an order of magnitude throughout the size spectra. -

Dental Insights Into Ophiuroids: Feeding Mechanisms and Life Style
Dental insights into ophiuroids: Feeding mechanisms and life style Karin Boos Biologische Anstalt Helgoland/ Alfred Wegener Institut for Polar and Marine Research, PO Box 180, 27483 Helgoland; [email protected] Ophiuroid echinoderms are highly specific towards different habitats reflecting lifestyles and feeding mechanisms. Previous studies have considered ophiuroids to be generally omnivorous macro- or microphageous feeders. According to their lifestyle, however, different feeding mechanisms may have evolved e.g. deposit feeding, filter feeding or predation. Most ophiuroids typically show more than one feeding mechanism along with their main feeding mode. In the present study, the morphology of teeth and associated papillae on individual jaw elements (see figure right) from ophiuroids performing different lifestyles (epibenthic, infaunal or epibenthic-cryptic) are compared and discussed in relation to reported feeding mechanisms and diets (for explanation on teeth and papillae see numbers in the pictures and adjacent text sections). Schematic overview of the ‚mouth‘ side of an ophiuroid (from Hayward and Ryland, 1996) MACROPHAGEOUS and a single jaw element. The epibethic: predators, scavengers and deposit feeders 3 Ophiura albida has three conical infradental papillae1 and two or three broadened 2 1 oral papillae located along 2 the lateral sides of the jaw plates. Long and strongly 3 pointed sharp teeth3 are found 3 down the vertical jaw edges. Epibenthic lifestyle of Ophiura albida Forbes, Top view of one jaw element in Ophiura Oblique view on the mouth in Ophiura Lateral view of jaws with teeth in Ophiura 1839; Photo by Encyclopedia of Marine Life albida. albida. albida. of Britain and Ireland MICROPHAGEOUS a) The infaunal: surface and sub-surface deposit feeders, filter feeder, suspension feeder 3 Amphiura filiformis and Acrocnida brachiata both 3 have a pair of slightly rounded infradental 1 1 1 1 papillae1, as well as two pairs of long and pointy 2 2 2 (A. -

Marlin Marine Information Network Information on the Species and Habitats Around the Coasts and Sea of the British Isles
MarLIN Marine Information Network Information on the species and habitats around the coasts and sea of the British Isles Amphiura filiformis and Ennucula tenuis in circalittoral and offshore sandy mud MarLIN – Marine Life Information Network Marine Evidence–based Sensitivity Assessment (MarESA) Review Eliane De-Bastos 2016-06-20 A report from: The Marine Life Information Network, Marine Biological Association of the United Kingdom. Please note. This MarESA report is a dated version of the online review. Please refer to the website for the most up-to-date version [https://www.marlin.ac.uk/habitats/detail/1107]. All terms and the MarESA methodology are outlined on the website (https://www.marlin.ac.uk) This review can be cited as: De-Bastos, E.S.R. 2016. [Amphiura filiformis] and [Ennucula tenuis] in circalittoral and offshore sandy mud. In Tyler-Walters H. and Hiscock K. (eds) Marine Life Information Network: Biology and Sensitivity Key Information Reviews, [on-line]. Plymouth: Marine Biological Association of the United Kingdom. DOI https://dx.doi.org/10.17031/marlinhab.1107.1 The information (TEXT ONLY) provided by the Marine Life Information Network (MarLIN) is licensed under a Creative Commons Attribution-Non-Commercial-Share Alike 2.0 UK: England & Wales License. Note that images and other media featured on this page are each governed by their own terms and conditions and they may or may not be available for reuse. Permissions beyond the scope of this license are available here. Based on a work at www.marlin.ac.uk (page left blank) -

Effects of Chronic Bottom Trawling on Soft Seafloor Macrofauna in The
Effects of chronic bottom trawling on soft seafloor macrofauna in the ANGOR UNIVERSITY Kattegat Skold, Mattias; Göransson, Peter; Jonsson, Patrik; Bastardie, Francois; Blomqvist, Mats; Agrenius, Stefan; Hiddink, Jan; Nilsson, Hans C.; Bartolino, Valerio Marine Ecology Progress Series DOI: 10.3354/meps12434 PRIFYSGOL BANGOR / B Published: 01/01/2018 Peer reviewed version Cyswllt i'r cyhoeddiad / Link to publication Dyfyniad o'r fersiwn a gyhoeddwyd / Citation for published version (APA): Skold, M., Göransson, P., Jonsson, P., Bastardie, F., Blomqvist, M., Agrenius, S., Hiddink, J., Nilsson, H. C., & Bartolino, V. (2018). Effects of chronic bottom trawling on soft seafloor macrofauna in the Kattegat. Marine Ecology Progress Series, 586, 41-55. https://doi.org/10.3354/meps12434 Hawliau Cyffredinol / General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. 01. Oct. 2021 1 Effects of chronic bottom trawling on soft seafloor macrofauna in the 2 Kattegat 3 4 Running page head: Effects of trawling on macrofauna in the Kattegat 5 6 Authors: Mattias Sköld1*, Peter Göransson2, Patrik Jonsson1, Francois Bastardie3, Mats 7 Blomqvist4, Stefan Agrenius5, Jan Geert Hiddink6, Hans C. -

A Puzzling Homology: a Brittle Star Using a Putative Cnidarian-Type
Downloaded from http://rsob.royalsocietypublishing.org/ on June 30, 2017 A puzzling homology: a brittle star using a putative cnidarian-type luciferase for rsob.royalsocietypublishing.org bioluminescence Je´roˆme Delroisse1, Esther Ullrich-Lu¨ter2, Stefanie Blaue2, Olga Ortega- Martinez3, Igor Eeckhaut1, Patrick Flammang1 and Je´roˆme Mallefet4 Research 1Research Institute for Biosciences, Biology of Marine Organisms and Biomimetics, University of Mons - UMONS, 23 Place du Parc, 7000 Mons, Belgium 2 Cite this article: Delroisse J, Ullrich-Lu¨ter E, Museum fu¨r Naturkunde, Leibniz Institute for Evolution and Biodiversity Science, Invalidenstr. 43, 10115 Berlin, Germany Blaue S, Ortega-Martinez O, Eeckhaut I, 3Department of Marine Science, The Sven Love´n Centre for Marine Sciences – Kristineberg, University of Flammang P, Mallefet J. 2017 A puzzling Gothenburg, 45178 Fiskeba¨ckskil, Sweden homology: a brittle star using a putative 4Marine Biology Laboratory, Universite´ Catholique de Louvain, ELI, 3 Place Croix du Sud L7.04.06, 1348 Louvain- cnidarian-type luciferase for bioluminescence. La-Neuve, Belgium Open Biol. 7: 160300. JD, 0000-0002-9233-6470 http://dx.doi.org/10.1098/rsob.160300 Bioluminescence relies on the oxidation of a luciferin substrate catalysed by a luciferase enzyme. Luciferins and luciferases are generic terms used to describe a large variety of substrates and enzymes. Whereas luciferins can Received: 30 October 2016 be shared by phylogenetically distant organisms which feed on organisms producing them, luciferases have been thought to be lineage-specific Accepted: 6 March 2017 enzymes. Numerous light emission systems would then have co-emerged independently along the tree of life resulting in a plethora of non-homologous luciferases. -

Global Diversity of Brittle Stars (Echinodermata: Ophiuroidea)
Review Global Diversity of Brittle Stars (Echinodermata: Ophiuroidea) Sabine Sto¨ hr1*, Timothy D. O’Hara2, Ben Thuy3 1 Department of Invertebrate Zoology, Swedish Museum of Natural History, Stockholm, Sweden, 2 Museum Victoria, Melbourne, Victoria, Australia, 3 Department of Geobiology, Geoscience Centre, University of Go¨ttingen, Go¨ttingen, Germany fossils has remained relatively low and constant since that date. Abstract: This review presents a comprehensive over- The use of isolated skeletal elements (see glossary below) as the view of the current status regarding the global diversity of taxonomic basis for ophiuroid palaeontology was systematically the echinoderm class Ophiuroidea, focussing on taxono- introduced in the early 1960s [5] and initiated a major increase in my and distribution patterns, with brief introduction to discoveries as it allowed for complete assemblages instead of their anatomy, biology, phylogeny, and palaeontological occasional findings to be assessed. history. A glossary of terms is provided. Species names This review provides an overview of global ophiuroid diversity and taxonomic decisions have been extracted from the literature and compiled in The World Ophiuroidea and distribution, including evolutionary and taxonomic history. It Database, part of the World Register of Marine Species was prompted by the near completion of the World Register of (WoRMS). Ophiuroidea, with 2064 known species, are the Marine Species (http://www.marinespecies.org) [6], of which the largest class of Echinodermata. A table presents 16 World Ophiuroidea Database (http://www.marinespecies.org/ families with numbers of genera and species. The largest ophiuroidea/index.php) is a part. A brief overview of ophiuroid are Amphiuridae (467), Ophiuridae (344 species) and anatomy and biology will be followed by a systematic and Ophiacanthidae (319 species).