Surveillance and Pathogen Characterization Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Report on Evaluation of Empowerment of Women in District Mansehra Through Women Friendly Halls
Report on Evaluation of Empowerment of Women in District Mansehra through Women Friendly Halls Sidra Fatima Minhas 11/27/2012 Table of Contents Executive Summary .............................................................................................................. 4 1. Women Freindly Halls (WFH) ......................................................................................... 5 1.1 Introduction ............................................................................................................. 8 1.1.1 Geographical Background ................................................................................ 9 1.1.2 Socio Cultural Context .....................................................................................12 1.1.3 Women Friendly Halls Project .........................................................................12 1.1.4 Objectives of WFHs Project ............................................................................13 1.2 Presence and Activities of Other Players ................................................................14 1.3 Rationale of the Evaluation .....................................................................................15 1.3.1 Objectives and Aim of the Evaluation ..............................................................15 1.4 Scope of the Evaluation .........................................................................................16 1.4.1 Period and Course of Evaluation .....................................................................16 1.4.2 Geographical -
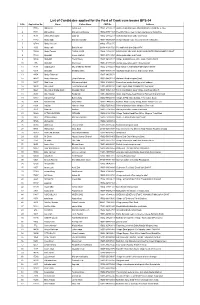
List of Candidates Applied for the Post of Cook-Cum-Bearer BPS-04 S.No Application No
List of Candidates applied for the Post of Cook-cum-bearer BPS-04 S.No Application No. Name Father Name CNIC No. Address 1 17715 Abbas Ali Akbar Gul 17301-2133451-1 Shaheen Muslim town, Mohallah Nazer Abad No 2, Hou 2 17171 Abbas khan Muhammad Sarwar 15602-0357410-5 The City School near to wali swat palace Saidu,Sha 3 7635 abbas khan zafar zafar ali 15602-2565421-7 mohalla haji abad saidu sharif swat 4 11483 Abdul aziz Muhammad jalal 15601-4595672-7 village baidara moh: school area teh:matta dist : 5 12789 Abdul Haleem - 90402-3773217-5 - 6 3090 Abdul Jalil Bakht Munir 21104-4193372-9 P/o and tehsil khar Bajur KPK 7 15909 Abdul Tawab FAZAL AHAD 15602-1353439-1 NAWAKALY MUHALA QANI ABADAN TEHSIL BABOZAY SWAT 8 17442 Abdullah Aleem bakhsh 15607-0374932-1 Afsar abad saidu sharif swat 9 10806 Abdullah Fazal Raziq 15607-0409351-5 Village Gulbandai p.o office saidu sharif tehsil b 10 255 Abdullah Mehmood 15602-6117033-5 Jawad super store panr mingora swat 11 7698 ABDULLAH MUHAMMAD IKRAM 15602-2135660-7 MOH BACHA AMANKOT MINGORA SWAT 12 4215 Abdullah Sarfaraz khan 15607-0359153-1 Almadina Model School and College Swat 13 4454 Abdur Rahman - 15607-0430767-1 - 14 14147 Abdur Rahman Umar Rahman 15602-0449712-7 Rahman Abad mingora Swat 15 12477 Abid khan Muhammad iqbal 15602-2165882-7 khunacham saidu sharif po tehsil babuxai 16 14428 Abu Bakar Tela Muhammad 17301-6936018-1 Ghari Inayat Abad Gulbahar # 2 Peshawar 17 10621 Abu Bakar Sadiq Shah Muqadar Shah 15607-0352136-7 Tehsil and District Swat Village and Post office R 18 9436 Abu Bakkar Barkat Ali 15607-0443898-5 Ikram Cap House Near Kashmir Backers Nishat Chowk 19 13113 Adnan Khan Bakht Zada 15602-9084073-3 Village & P/O: Bara Bandai, Teh: Kabal ,Swat. -

Khyber Pakhtunkhwa - Daily Flood Report Date (29 09 2011)
Khyber Pakhtunkhwa - Daily Flood Report Date (29 09 2011) SWAT RIVER Boundary 14000 Out Flow (Cusecs) 12000 International 10000 8000 1 3 5 Provincial/FATA 6000 2 1 0 8 7 0 4000 7 2 4 0 0 2 0 3 6 2000 5 District/Agency 4 4 Chitral 0 Gilgit-Baltistan )" Gauge Location r ive Swat River l R itra Ch Kabul River Indus River KABUL RIVER 12000 Khyber Pakhtunkhwa Kurram River 10000 Out Flow (Cusecs) Kohistan 8000 Swat 0 Dir Upper Nelam River 0 0 Afghanistan 6000 r 2 0 e 0 v 0 i 1 9 4000 4 6 0 R # 9 9 5 2 2 3 6 a Dam r 3 1 3 7 0 7 3 2000 o 0 0 4 3 7 3 1 1 1 k j n ") $1 0 a Headworks P r e iv Shangla Dir L")ower R t a ¥ Barrage w Battagram S " Man")sehra Lake ") r $1 Amandara e v Palai i R Malakand # r r i e a n Buner iv h J a R n ") i p n Munda n l a u Disputed Areas a r d i S K i K ") K INDUS RIVER $1 h Mardan ia ") ") 100000 li ") Warsak Adezai ") Tarbela Out Flow (Cusecs) ") 80000 ") C")harsada # ") # Map Doc Name: 0 Naguman ") ") Swabi Abbottabad 60000 0 0 Budni ") Haripur iMMAP_PAK_KP Daily Flood Report_v01_29092011 0 0 ") 2 #Ghazi 1 40000 3 Peshawar Kabal River 9 ") r 5 wa 0 0 7 4 7 Kh 6 7 1 6 a 20000 ar Nowshera ") Khanpur r Creation Date: 29-09-2011 6 4 5 4 5 B e Riv AJK ro Projection/Datum: GCS_WGS_1984/ D_WGS_1984 0 Ghazi 2 ") #Ha # Web Resources: http://www.immap.org Isamabad Nominal Scale at A4 paper size: 1:3,500,000 #") FATA r 0 25 50 100 Kilometers Tanda e iv Kohat Kohat Toi R s Hangu u d ") In K ai Map data source(s): tu Riv ") er Punjab Hydrology Irrigation Division Peshawar Gov: KP Kurram Garhi Karak Flood Cell , UNOCHA RIVER $1") Baran " Disclaimers: KURRAM RIVER G a m ") The designations employed and the presentation of b e ¥ Kalabagh 600 Bannu la material on this map do not imply the expression of any R K Out Flow (Cusecs) iv u e r opinion whatsoever on the part of the NDMA, PDMA or r ra m iMMAP concerning the legal status of any country, R ") iv ") e K territory, city or area or of its authorities, or concerning 400 r h ") ia the delimitation of its frontiers or boundaries. -

Ethnobotanical Survey of the Flora of Tehsil Balakot, District Mansehra, Khyber Pakhtunkhwa, Pakistan
Science Arena Publications Specialty Journal of Biological Sciences ISSN: 2412-7396 Available online at www.sciarena.com 2018, Vol, 4 (3): 7-14 Ethnobotanical Survey of the Flora of Tehsil Balakot, District Mansehra, Khyber Pakhtunkhwa, Pakistan Muhammad Irfan1, 2*, Danish Ali3, Gul Jan1, Waheed Murad1 1Department of Botany, Abdulwalikhan University, Mardan, Pakistan, 2Department of Botany, University of Swabi, Swabi, Pakistan, 3Department of Botany, Government Post Graduate College, Mansehra, Pakistan. *Corresponding Author Abstract : The inhabitants of tehsil Balakot, district Mansehra, Khyber Pakhtunkhwa, Pakistan were using eighty-eight different taxa belonging to seventy-one genera and forty-six families. Amongst the parts used, the results showed that the whole plant of most of them viz. thirty-two, the leaves of twenty-seven, the stem of twelve, and the fruits of nine taxa were used for different purposes. The governing families of the study area were Rosaceae Family consisting of thirteen taxa, solanaceae having five taxa, Buxaceae, Verbenaceae, Lamiaceae, Acanthaceae having three taxa each, Papilionaceae, Berberidaceae, Buddlejaceae, Anacardiaceae, Thymelaceae, Sapindaceae, Araliaceae, Guttiferae, Oleaceae, Cupressaceae, Euphorbiaceae, Poaceae, Rhamnaceae, Rutaceae, Caprifoliaceae keeping two taxa, and the rest of all the families contained one taxa, each was recorded in the study area. Keywords: Ethnobotanical, Medicinal, Balakot, Mansehra, Khyber Pakhtunkhwa, Pakistan. INTRODUCTION Balakot is located between 34°33′N 73°21′E latitude and 34°33′N 73°21′E longitude near Kaghan valley located in the Northern part of Pakistan. It is a historical place and famous tourism site of the region, and the gateway to Kaghan valley of Khyber Pakhtunkhwa, Pakistan. The famous river of Balakot is Kunhar, originating from Lulusar lake and merges with River Jehlum just outside Muzaffarabad in Azad Kashmir. -

Download Map (PDF | 1.07
Palas Palas Palas Kohistan Kel Allai Mansehra Alpuri Kaghan Sharda v Guraze Batagram Puran Bala Kot Athmuqam Devli Jaberr Neelum P. A . K Kewal Bassi Khel Dundnial Batagram v Shangla Mohandri Hilkot Sacha Kalan Hangrai Jaborr Kundal Shahi Chattar Plain Ghanool Nusrat Khel Bhogarmong Shahkot Battal F.R.Kala Dhaka Satbani Neelum Dilborri Icherrian Akazai Bheri Talgran Garlat Shamdarra Machyara Belian Sum Ellahi Mong Saidpur Hassan Khel Tanda Ashkot Shaukat Abad Balakot Badl Shungli Sirli Sacha Shinkiari Kahori Kathal Ogai Mansehra Inayat Abad Shohal Mazullah Oghi Balgran Trangri Sabir Shah Baffa Town Heer Kutli N.W.F.P Dhodial Talhata Shergarh Malik Pur Barian Nika Pani Mada Khel Noora Seri Punjfran Challiana Attar Shisha Garhi Habibullah Karorri Bherkund Karnol Road blocked due to landslide Perhinna Hamsherian Sandasar Muzaffarabad Punjkot Mansehra City No.2 Laber Kot Darband Gojra !( 2 Houses Damaged Mansehra City No.1 Pairan Muzaffarabad Punjkot 3 Persons injured Phulrra Mansehra City No.3 Mansehra Rural Muzaffarabad Jaloo Chattar Domel Dara Shanaya Datta Banamoola Langerpura Leepa Swan Miara km 10 50 km 50 20 km 20 40 km 40 Behall Charakpura Hattian Dopatta km 30 Lassan Thkral 5.4 Lassan Nawab (! Tharian Kai Manja Lamnian Garhi Dopatta Hattian DISPUTED AREA Kot Komi Chak Hama Chatter Kalass Hattian Langla BHU Chinari Jhand Gran Hattian Bala Chikarv Danna Gujar Bandi RHC Chakothi Mera Kalan Sena Daman Abbottabad Kacheli Salmia Chinari v Hill Surang v Katkair Chamyati Bir Pani Khalana Dhir Kot Rangla Thub Sahlian vDhoundan FANA Mallot -

RFP Document 11-12-2020.Pdf
Utility Stores Corporation (USC) Tender Document For Supply, Installation, Integration, Testing, Commissioning & Training of Next Generation Point of Sale System as Lot-1 And End-to-end Data Connectivity along with Platform Hosting Services as Lot-2 Of Utility Stores Locations Nationwide on Turnkey Basis Date of Issue: December 11, 2020 (Friday) Date of Submission: December 29, 2020 (Tuesday) Utility Stores Corporation of Pakistan (Pvt) Ltd, Head Office, Plot No. 2039, F-7/G-7 Jinnah Avenue, Blue Area, Islamabad Phone: 051-9245047 www.usc.org.pk Page 1 of 18 TABLE OF CONTENTS 1. Introduction ....................................................................................................................... 3 2. Invitation to Bid ................................................................................................................ 3 3. Instructions to Bidders ...................................................................................................... 4 4. Definitions ......................................................................................................................... 5 5. Interpretations.................................................................................................................... 7 6. Headings & Tiles ............................................................................................................... 7 7. Notice ................................................................................................................................ 7 8. Tender Scope .................................................................................................................... -

Punjab Region & Islamabad Offices
Punjab Region & Islamabad Offices Regioanl/Divisioanl/Tehsil office Contact No. S.No Name Complete Address Regional Office Punjab & 48-L, Model Town Extension, Lahore 04235219116-18 1 Divisional office Lahore 2 Divisional Office H.No.805, St. No.1, Chaklala Scheme-051-9281132 III, Rawalpindi, District Rawalpindi Rawalpindi H.No.805, St. No.1, Chaklala Scheme-051-9281130 3 III, Rawalpindi, District Rawalpindi 4 Gujar Khan House no. D-37,Scheem-I,main gt 051-3516533 road,gujarkhan Kahuta H. No. A-003, Kahuta, District 051-3311256 5 Rawalpindi 6 Kallar Syedan Qasam Market,Pindi Road, Kallar 051-3570157 Sayaddan, District Rawalpindi Kotli Sattian Police Station Road, Kotli Sattian, 051-3356383 7 District Rawalpindi 8 Murree Ijaz Abbasi Flats, Walker Estate, 051-3756642 Sunny bank (near Aara Machine), Taxila House no B-27,Wahadat 051-4254007 9 colony,thana taxila,taxila 10 Attock H.# C12,Block-A,Peoples Colony 057-2541354 ,Attock Hazro Hattian Road Bhadur Khan Stop 057-2642251 P/O Mossa Mohalla Khalil Pura Shahid Market Jamia Masjid Gulzar- e-Habib Hazro, District Attock 11 12 Fateh Jang Fateh Jang, District Attock 057-2210665 Jand Mohalla Aminabad, Ward No. 1, 057-2621004 13 Kohat Raod, Jand, District Attock 14 Pindigheb H. No. B-XI-292 C/1, Zafar Chowk, 057-2352250 Pindi Gheb, District Attock Hasanabdal House No B 111-317 1st floor hazara 057-2523494 15 road Hasanabdal 16 Chakwal H. No. MCB-3/440, Mohalla 0543-550224 Saddique Abad, Line Park, District Chakwal Kallar Kahar Sargodha Road, Near TMA 0543-588110 Building, Kallar Kahar, District 17 Chakwal 18 Choa Saidan Shah Choa Saidan Shah, District Chakwal 0543-580648 Talagang H. -

Contesting Candidates NA-1 Peshawar-I
Form-V: List of Contesting Candidates NA-1 Peshawar-I Serial No Name of contestng candidate in Address of contesting candidate Symbol Urdu Alphbeticl order Allotted 1 Sahibzada PO Ashrafia Colony, Mohala Afghan Cow Colony, Peshawar Akram Khan 2 H # 3/2, Mohala Raza Shah Shaheed Road, Lantern Bilour House, Peshawar Alhaj Ghulam Ahmad Bilour 3 Shangar PO Bara, Tehsil Bara, Khyber Agency, Kite Presented at Moh. Gul Abad, Bazid Khel, PO Bashir Ahmad Afridi Badh Ber, Distt Peshawar 4 Shaheen Muslim Town, Peshawar Suitcase Pir Abdur Rehman 5 Karim Pura, H # 282-B/20, St 2, Sheikhabad 2, Chiragh Peshawar (Lamp) Jan Alam Khan Paracha 6 H # 1960, Mohala Usman Street Warsak Road, Book Peshawar Haji Shah Nawaz 7 Fazal Haq Baba Yakatoot, PO Chowk Yadgar, H Ladder !"#$%&'() # 1413, Peshawar Hazrat Muhammad alias Babo Maavia 8 Outside Lahore Gate PO Karim Pura, Peshawar BUS *!+,.-/01!234 Khalid Tanveer Rohela Advocate 9 Inside Yakatoot, PO Chowk Yadgar, H # 1371, Key 5 67'8 Peshawar Syed Muhammad Sibtain Taj Agha 10 H # 070, Mohala Afghan Colony, Peshawar Scale 9 Shabir Ahmad Khan 11 Chamkani, Gulbahar Colony 2, Peshawar Umbrella :;< Tariq Saeed 12 Rehman Housing Society, Warsak Road, Fist 8= Kababiyan, Peshawar Amir Syed Monday, April 22, 2013 6:00:18 PM Contesting candidates Page 1 of 176 13 Outside Lahori Gate, Gulbahar Road, H # 245, Tap >?@A= Mohala Sheikh Abad 1, Peshawar Aamir Shehzad Hashmi 14 2 Zaman Park Zaman, Lahore Bat B Imran Khan 15 Shadman Colony # 3, Panal House, PO Warsad Tiger CDE' Road, Peshawar Muhammad Afzal Khan Panyala 16 House # 70/B, Street 2,Gulbahar#1,PO Arrow FGH!I' Gulbahar, Peshawar Muhammad Zulfiqar Afghani 17 Inside Asiya Gate, Moh. -

Shamlai District Batagram, Mohandri Tehsil Balakot) October 2007 to May 2008
Federal Department of Foreign Affairs FDFA Swiss Agency for Development and Cooperation SDC Swiss Humanitarian Aid and SHA HUMANITARIAN AID (SDC-HA) PAKISTAN RECONSTRUCTION AND LIVELIHOOD PROGRAMME RLP Housing Reconstruction Programme Pakistan Report on Model Houses in the two union councils (Shamlai District Batagram, Mohandri Tehsil Balakot) October 2007 to May 2008 Authors: Muhammad Yousuf HRC Engineer Batagram Asad Farooq Head of HRC Balakot Contributor: Martin Weiersmueller Programme Advisor Reconstruction and Livelihood Programme (RLP) Azam Beg Tajik Programme Manager Community Development Mansehra 16th June 2008 Table of Contents 1 Background and need of model houses ........................................................................................................... 3 1.1 Procedure for selection of model house.................................................................................................. 3 1.2 Out comes of model house ..................................................................................................................... 3 2 Technical aspects of model house ................................................................................................................... 4 2.1 Types of model house............................................................................................................................. 4 2.1.1 Dhajji model house........................................................................................................................ 4 2.1.2 Technical specification -

(Female) Mansehra
OFFICE OF THE DISTRICT EDUCATION OFFICER (FEMALE) MANSEHRA STATEMENT SHOWING THE VACANT POST OF DISTRICT CADRES S# EMIS Code Name of School Union Council Pst Grand Total 1 37523 GGPS Charach Behali 1 1 2 41084 GGPS Kanait Hamsharian 1 1 3 18383 GGPS Battal Payeen Lassan Nawab 1 1 4 18647 GGPS Sherpur Malik Pur 1 1 5 18654 GGPS Susal Gawon Malik Pur 1 1 6 18439 GGPS Ganda Mansehra Deh 1 1 7 18437 GGPS Fojdara Parhinna 1 1 8 36758 GGPS Tarari Phulra 1 1 9 18563 GGPS Mohar Kalan Sawan Maira 1 1 10 NCP GGPS Javer Bandi Shungli 3 3 11 40909 GGPS Kolika Belian 2 2 12 18442 GGPS Ghanian Dilbori 4 4 13 32354 GGPS Sheikh Abad Shamdhara 2 2 14 18338 GGPS Baffa Battagram BAFFA 1 1 15 32288 GGPS METHRA JALGALI BATTAL 1 1 16 NYA GGPS Bagroo HILKOT 4 4 17 32377 GGPS Jaber Gali JABAR DEVLI 3 3 18 32245 GGPS Batkarar Hassa GARLAT 1 1 19 Newly EstablishGGPS Khalian GHANOOL 3 3 20 18621 GGPS Sandoo Sangar GHANOOL 2 2 21 38720 GGPS AGLA GRAN KAGHAN 2 2 22 Newly EstablishGGPS Chowki Khair Abad KARNOL 3 3 23 Newly EstablishGGPS SAYL KARNOL 3 3 24 18640 GGPS SHALI JARED MOHANDRI 1 1 25 18568 GGPS More Baffa Kalan Lassan Thakral 1 1 26 39236 GGPS Taradda Shoukat Abad 1 1 27 18487 GGPS Kathai Kathai 2 2 28 32300 GGPS Ramkot Shergarh 1 1 29 29210 GGPS MUHALLAH M.SHAH CHATTAR PLAIN 2 2 30 18614 GGPS Sabir Shah TRANGRI SABIR SHAH 2 2 31 18660 GGPS Suwan HUNGRAI 1 1 32 32349 GGPS Kagal Tallahatta 1 1 33 39184 GGPs Angar Ban Nika Pani 1 1 34 18517 GGPS Khandgran Shanaya Bala 1 1 35 18449 GGPS Ghotar Attershisha 1 1 36 18636 GGPS Shakot Potha Behali 1 1 37 32770 GGPS -

DFG Part-L Development Settled
DEMANDS FOR GRANTS DEVELOPMENTAL EXPENDITURE FOR 2020–21 VOL-III (PART-L) GOVERNMENT OF KHYBER PAKHTUNKHWA FINANCE DEPARTMENT REFERENCE TO PAGES DFG PART- L GRANT # GRANT NAME PAGE # - SUMMARY 01 – 23 50 DEVELOPMENT 24 – 177 51 RURAL AND URBAN DEVELOPMENT 178 – 228 52 PUBLIC HEALTH ENGINEERING 229 – 246 53 EDUCATION AND TRAINING 247 – 291 54 HEALTH SERVICES 292 – 337 55 CONSTRUCTION OF IRRIGATION 338 – 385 CONSTRUCTION OF ROADS, 56 386 – 456 HIGHWAYS AND BRIDGES 57 SPECIAL PROGRAMME 457 – 475 58 DISTRICT PROGRAMME 476 59 FOREIGN AIDED PROJECTS 477 – 519 ( i ) GENERAL ABSTRACT OF DISBURSEMENT (SETTLED) BUDGET REVISED BUDGET DEMAND MAJOR HEADS ESTIMATES ESTIMATES ESTIMATES NO. -

Saresh V Alley Seran V Alley Jabori
73.2 73.3 73.4 73.5 73.6 73.7 73.8 BATAGRAM DIST BTA NAH GUN ! ! ! Rajwal Bala ALLAI TEHS Gangwal GAN! alley Rajwal MANSEHRA DIST lla i V BALA KOT TEHS A Kinari 34.8 34.8 KA2 Kaghan ! ! KA1 MSP Pattan Dos No.2 ! Pattan Dos No.1 BLS ! BATAGRAM TEHS Seri Tarli Phagal Gul Dheri PHA ! Upper Kaghan Valley Badal Giran ! NZI Sharan Valley JBA ! Bela RON! ! ! 34.7 KHO Khola Mahandri Takia Bichhla ATHMUQAM TEHS 34.7 Chan Garhi Muhandari Joshal MAH Jabar Gali ! Deoli JAR ! ! DEV SHR Jarid Jabar BSS ey! SAS MAG ! ll! ! Bela ! a JCH ! BEL PAR Ochari ! ! ! V Interagency (ACTED)")! AUL ri ! Saeedabad !E Heli Landing Zone o SHO NAW ! b ! Bu ") Storage Site Adai Balimang a h J Hangrai n Prov. or Dist. Capital Sundi KAN KAW ! j ! DUB ! ! ORC a DHU SAD KAS Masach Kawai BatgranTehsil Capital Balimang ! Ban SEB !V ! ! Kayan Bhunja JagranUnion Council Capital Thanwan Dola a Sacha Kalan Banda ! Khola Malo Karah ! l TUN Sangra l AND Sig. Settlement Pamaroy Paprang BHSe y ! Hilkot JAB Methal ! y ! Kair Sachha JIG e GHE Ghanool Khass BOO ! e Kutgali ! Karakoram Hwy JUC ! l ! HAN ! l l NEELUM DIST Malkan Gali Gadda Seri Kamashian ! Chehla Jiggan l 34.6 Dabrian Bhatian Primary Route 34.6 ! a ! Rin BAA BRB a Mang Secondary Route BAS Trar ! V Batal ! ! Mandi MNG Tertiary Rt / Street Churian Dandar ! Bakki V ! SAN Qazi Khati Baki MRB! ! ! h Walking Track ! ! Khait Sarash Sangarh Seriari DBA Bhogarmang KUN s Gohar Bandi n SRH ! ! e Unverified BS1 r Patlang ARH NPA BS2 a ! ! Jal Gali ! ! Satbani ! MUZAFFARABAD DIST District Boundary! a Bhangian PLB r KUT Ahall DHD ! BAL ! Tehsil Boundary e Mitti Kot ! ! MUZAFFARABAD TEHS ! S KHN Bat Sanghar Serian CBA ! DA2 ! Dader Jadeed Bala Kot Garat PTT Ghana Ghumat Union C.