The Human Respiratory System and Its Microbiome at a Glimpse
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Gut Microbiota and Inflammation
International Journal of Environmental Research and Public Health Review The Gut Microbiota and Inflammation: An Overview 1, 2 1, 1, , Zahraa Al Bander *, Marloes Dekker Nitert , Aya Mousa y and Negar Naderpoor * y 1 Monash Centre for Health Research and Implementation, School of Public Health and Preventive Medicine, Monash University, Melbourne 3168, Australia; [email protected] 2 School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane 4072, Australia; [email protected] * Correspondence: [email protected] (Z.A.B.); [email protected] (N.N.); Tel.: +61-38-572-2896 (N.N.) These authors contributed equally to this work. y Received: 10 September 2020; Accepted: 15 October 2020; Published: 19 October 2020 Abstract: The gut microbiota encompasses a diverse community of bacteria that carry out various functions influencing the overall health of the host. These comprise nutrient metabolism, immune system regulation and natural defence against infection. The presence of certain bacteria is associated with inflammatory molecules that may bring about inflammation in various body tissues. Inflammation underlies many chronic multisystem conditions including obesity, atherosclerosis, type 2 diabetes mellitus and inflammatory bowel disease. Inflammation may be triggered by structural components of the bacteria which can result in a cascade of inflammatory pathways involving interleukins and other cytokines. Similarly, by-products of metabolic processes in bacteria, including some short-chain fatty acids, can play a role in inhibiting inflammatory processes. In this review, we aimed to provide an overview of the relationship between the gut microbiota and inflammatory molecules and to highlight relevant knowledge gaps in this field. -

Case 16-2019: a 53-Year-Old Man with Cough and Eosinophilia
The new england journal of medicine Case Records of the Massachusetts General Hospital Founded by Richard C. Cabot Eric S. Rosenberg, M.D., Editor Virginia M. Pierce, M.D., David M. Dudzinski, M.D., Meridale V. Baggett, M.D., Dennis C. Sgroi, M.D., Jo-Anne O. Shepard, M.D., Associate Editors Alyssa Y. Castillo, M.D., Case Records Editorial Fellow Emily K. McDonald, Sally H. Ebeling, Production Editors Case 16-2019: A 53-Year-Old Man with Cough and Eosinophilia Rachel P. Simmons, M.D., David M. Dudzinski, M.D., Jo-Anne O. Shepard, M.D., Rocio M. Hurtado, M.D., and K.C. Coffey, M.D. Presentation of Case From the Department of Medicine, Bos- Dr. David M. Dudzinski: A 53-year-old man was evaluated in an urgent care clinic of ton Medical Center (R.P.S.), the Depart- this hospital for 3 months of cough. ment of Medicine, Boston University School of Medicine (R.P.S.), the Depart- Five years before the current evaluation, the patient began to have exertional ments of Medicine (D.M.D., R.M.H.), dyspnea and received a diagnosis of hypertrophic obstructive cardiomyopathy, with Radiology (J.-A.O.S.), and Pathology a resting left ventricular outflow gradient of 110 mm Hg on echocardiography. (K.C.C.), Massachusetts General Hos- pital, and the Departments of Medicine Although he received medical therapy, symptoms persisted, and percutaneous (D.M.D., R.M.H.), Radiology (J.-A.O.S.), alcohol septal ablation was performed 1 year before the current evaluation, with and Pathology (K.C.C.), Harvard Medical resolution of the exertional dyspnea. -

Infections of the Respiratory Tract
F70954-07.qxd 12/10/02 7:36 AM Page 71 Infections of the respiratory 7 tract the nasal hairs and by inertial impaction with mucus- 7.1 Pathogenesis 71 covered surfaces in the posterior nasopharynx (Fig. 11). 7.2 Diagnosis 72 The epiglottis, its closure reflex and the cough reflex all reduce the risk of microorganisms reaching the lower 7.3 Management 72 respiratory tract. Particles small enough to reach the tra- 7.4 Diseases and syndromes 73 chea and bronchi stick to the respiratory mucus lining their walls and are propelled towards the oropharynx 7.5 Organisms 79 by the action of cilia (the ‘mucociliary escalator’). Self-assessment: questions 80 Antimicrobial factors present in respiratory secretions further disable inhaled microorganisms. They include Self-assessment: answers 83 lysozyme, lactoferrin and secretory IgA. Particles in the size range 5–10 µm may penetrate further into the lungs and even reach the alveolar air Overview spaces. Here, alveolar macrophages are available to phagocytose potential pathogens, and if these are overwhelmed neutrophils can be recruited via the This chapter deals with infections of structures that constitute inflammatory response. The defences of the respira- the upper and lower respiratory tract. The general population tory tract are a reflection of its vulnerability to micro- commonly experiences upper respiratory tract infections, bial attack. Acquisition of microbial pathogens is which are often seen in general practice. Lower respiratory tract infections are less common but are more likely to cause serious illness and death. Diagnosis and specific chemotherapy of respiratory tract infections present a particular challenge to both the clinician and the laboratory staff. -

Care Process Models Streptococcal Pharyngitis
Care Process Model MONTH MARCH 20152019 DEVELOPMENTDIAGNOSIS AND AND MANAGEMENT DESIGN OF OF CareStreptococcal Process Models Pharyngitis 20192015 Update This care process model (CPM) was developed by Intermountain Healthcare’s Antibiotic Stewardship team, Medical Speciality Clinical Program,Community-Based Care, and Intermountain Pediatrics. Based on expert opinion and the Infectious Disease Society of America (IDSA) Clinical Practice Guidelines, it provides best-practice recommendations for diagnosis and management of group A streptococcal pharyngitis (strep) including the appropriate use of antibiotics. WHAT’S INSIDE? KEY POINTS ALGORITHM 1: DIAGNOSIS AND TREATMENT OF PEDIATRIC • Accurate diagnosis and appropriate treatment can prevent serious STREPTOCOCCAL PHARYNGITIS complications . When strep is present, appropriate antibiotics can prevent AGES 3 – 18 . 2 SHU acute rheumatic fever, peritonsillar abscess, and other invasive infections. ALGORITHM 2: DIAGNOSIS Treatment also decreases spread of infection and improves clinical AND TREATMENT OF ADULT symptoms and signs for the patient. STREPTOCOCCAL PHARYNGITIS . 4 • Differentiating between a patient with an active strep infection PHARYNGEAL CARRIERS . 6 and a patient who is a strep carrier with an active viral pharyngitis RESOURCES AND REFERENCES . 7 is challenging . Treating patients for active strep infection when they are only carriers can result in overuse of antibiotics. Approximately 20% of asymptomatic school-aged children may be strep carriers, and a throat culture during a viral illness may yield positive results, but not require antibiotic treatment. SHU Prescribing repeat antibiotics will not help these patients and can MEASUREMENT & GOALS contribute to antibiotic resistance. • Ensure appropriate use of throat • For adult patients, routine overnight cultures after a negative rapid culture for adult patients who meet high risk criteria strep test are unnecessary in usual circumstances because the risk for acute rheumatic fever is exceptionally low. -

( 12 ) United States Patent
US010314866B2 (12 ) United States Patent (10 ) Patent No. : US 10 ,314 ,866 B2 Kovarik ( 45 ) Date of Patent: * Jun . 11, 2019 ( 54 ) METHOD OF REDUCING THE 61/ 919 , 297 , filed on Dec . 20 , 2013, provisional LIKELIHOOD OF SKIN CANCER IN AN application No . 61/ 467, 767 , filed on Mar . 25, 2011 . INDIVIDUAL HUMAN BEING (71 ) Applicant: Joseph E . Kovarik , Englewood , CO (51 ) Int. Cl. (US ) A61K 31 / 58 ( 2006 . 01 ) A61K 35 /00 (2006 . 01 ) (72 ) Inventor: Joseph E . Kovarik , Englewood , CO A61K 35 / 74 (2015 . 01 ) ( US ) A61K 38 / 17 ( 2006 .01 ) A61K 31 / 715 ( 2006 . 01 ) ( * ) Notice : Subject to any disclaimer , the term of this patent is extended or adjusted under 35 (32 ) U . S . Cl. CPC .. .. .. .. A61K 35 / 74 ( 2013 .01 ) ; A61K 31 /58 U . S . C . 154 ( b ) by 0 days . (2013 .01 ) ; A61K 31/ 715 (2013 . 01 ) ; A61K This patent is subject to a terminal dis 38 / 1709 (2013 . 01 ) ; A61K 38 / 1758 ( 2013 .01 ) ; claimer . A61K 2035 / 11 ( 2013 .01 ) (58 ) Field of Classification Search ( 21) Appl. No .: 16 / 160, 336 None (22 ) Filed : Oct . 15, 2018 See application file for complete search history . (65 ) Prior Publication Data ( 56 ) References Cited US 2019 / 0038680 A1 Feb . 7 , 2019 U . S . PATENT DOCUMENTS Related U . S . Application Data 3 , 178 , 341 A 4 / 1965 Hamill et al . 4 , 568 ,639 A 2 / 1986 Lew (63 ) Continuation of application No . 15 / 403 , 823 , filed on 4 ,687 , 841 A 8 / 1987 Spilburg et al. Jan . 11 , 2017 , now Pat. No. 10 , 111, 913 , which is a 4 , 720 ,486 A 1 / 1988 Spilburg et al . -

Studies on the Role of the Keratinocytes in Cutaneous Immnity
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repository of the Academy's Library Factors shaping the composition of the cutaneous microbiota K. Szabó1, L. Erdei2, B. Sz. Bolla2, G. Tax2, T. Bíró3, L. Kemény1,2 1. MTA-SZTE Dermatological Research Group, Szeged, Hungary 2. Department of Dermatology and Allergology, University of Szeged, Hungary 3. DE-MTA “Lendület” Cellular Physiology Research Group, Departments of Physiology and Immunology, Faculty of Medicine, University of Debrecen, Debrecen, Hungary Running head: Factors shaping the composition of the cutaneous microbiota Manuscript word count: Manuscript table count: none Manuscript figure count: none Corresponding author: Kornélia Szabó Tel: +36-62-545 799 Fax: +36-62-545 799 E-mail address: [email protected] Keywords: microbiota, cutaneous microbiota, Propionibacterium acnes, acne vulgaris, disappearing microbiota hypothesis What's already known about this topic: -Microbes are integral components of the human ecosystem. -The cutaneous microbiota plays an important role in the regulation of skin homeostasis. -The composition of skin microbiota is influenced by many factors. What does this study add? -The dominance of P. acnes in the postadolescent sebum-rich skin regions and its role in acne pathogenesis may be explained by the disappearing microbiota hypothesis. Funding sources: Hungarian Scientific Research Fund (OTKA NK105369), János Bolyai Research Scholarship from the Hungarian Academy of Sciences (for K. Sz). Conflict of interest: The authors declare no conflict of interest. 1 Abstract From our birth, we are constantly exposed to bacteria, fungi and viruses, some of which are capable of transiently or permanently inhabiting our different body parts as our microbiota. -

Diagnosis and Treatment of Acute Pharyngitis/Tonsillitis: a Preliminary Observational Study in General Medicine
Eur opean Rev iew for Med ical and Pharmacol ogical Sci ences 2016; 20: 4950-4954 Diagnosis and treatment of acute pharyngitis/tonsillitis: a preliminary observational study in General Medicine F. DI MUZIO, M. BARUCCO, F. GUERRIERO Azienda Sanitaria Locale Roma 4, Rome, Italy Abstract. – OBJECTIVE : According to re - pharmaceutical expenditure, without neglecting cent observations, the insufficiently targeted the more important and correct application of use of antibiotics is creating increasingly resis - the Guidelines with performing of a clinically val - tant bacterial strains. In this context, it seems idated test that carries advantages for reducing increasingly clear the need to resort to extreme the use of unnecessary and potentially harmful and prudent rationalization of antibiotic thera - antibiotics and the consequent lower prevalence py, especially by the physicians working in pri - and incidence of antibiotic-resistant bacterial mary care units. In clinical practice, actually the strains. general practitioner often treats multiple dis - eases without having the proper equipment. In Key Words: particular, the use of a dedicated, easy to use Acute pharyngitis, Tonsillitis, Strep throat, Beta-he - diagnostic test would be one more weapon for molytic streptococcus Group A (GABHS), Rapid anti - the correct diagnosis and treatment of acute gen detection test, Appropriateness use of antibiotics, pharyngo-tonsillitis. The disease is a condition Cost savings in pharmaceutical spending. frequently encountered in clinical practice but -

Common Questions About Streptococcal Pharyngitis MONICA G
Common Questions About Streptococcal Pharyngitis MONICA G. KALRA, DO, Memorial Family Medicine Residency, Sugar Land, Texas KIM E. HIGGINS, DO, Envoy Hospice and Brookdale Hospice, Fort Worth, Texas EVAN D. PEREZ, MD, Memorial Family Medicine Residency, Sugar Land, Texas Group A beta-hemolytic streptococcal (GABHS) infection causes 15% to 30% of sore throats in children and 5% to 15% in adults, and is more common in the late winter and early spring. The strongest independent predictors of GABHS pharyngitis are patient age of five to 15 years, absence of cough, tender anterior cervical adenopa- thy, tonsillar exudates, and fever. To diagnose GABHS pharyngitis, a rapid antigen detection test should be ordered in patients with a modified Centor or FeverPAIN score of 2 or 3. First-line treatment for GABHS pharyngitis includes a 10-day course of penicillin or amoxicillin. Patients allergic to penicillin can be treated with first- generation cephalosporins, clindamycin, or macrolide antibiotics. Nonsteroidal anti-inflammatory drugs are more effective than acet- aminophen and placebo for treatment of fever and pain associated with GABHS pharyngitis; medicated throat lozenges used every two hours are also effective. Corticosteroids provide only a small reduc- tion in the duration of symptoms and should not be used routinely. (Am Fam Physician. 2016;94(1):24-31. Copyright © 2016 American Academy of Family Physicians.) ILLUSTRATION JOHN BY KARAPELOU CME This clinical content haryngitis is diagnosed in 11 mil- EVIDENCE SUMMARY conforms to AAFP criteria lion persons in the outpatient set- Several risk factors should increase the index for continuing medical 1 education (CME). See ting each year in the United States. -

Downloaded from 3
Philips et al. BMC Genomics (2020) 21:402 https://doi.org/10.1186/s12864-020-06810-9 RESEARCH ARTICLE Open Access Analysis of oral microbiome from fossil human remains revealed the significant differences in virulence factors of modern and ancient Tannerella forsythia Anna Philips1, Ireneusz Stolarek1, Luiza Handschuh1, Katarzyna Nowis1, Anna Juras2, Dawid Trzciński2, Wioletta Nowaczewska3, Anna Wrzesińska4, Jan Potempa5,6 and Marek Figlerowicz1,7* Abstract Background: Recent advances in the next-generation sequencing (NGS) allowed the metagenomic analyses of DNA from many different environments and sources, including thousands of years old skeletal remains. It has been shown that most of the DNA extracted from ancient samples is microbial. There are several reports demonstrating that the considerable fraction of extracted DNA belonged to the bacteria accompanying the studied individuals before their death. Results: In this study we scanned 344 microbiomes from 1000- and 2000- year-old human teeth. The datasets originated from our previous studies on human ancient DNA (aDNA) and on microbial DNA accompanying human remains. We previously noticed that in many samples infection-related species have been identified, among them Tannerella forsythia, one of the most prevalent oral human pathogens. Samples containing sufficient amount of T. forsythia aDNA for a complete genome assembly were selected for thorough analyses. We confirmed that the T. forsythia-containing samples have higher amounts of the periodontitis-associated species than the control samples. Despites, other pathogens-derived aDNA was found in the tested samples it was too fragmented and damaged to allow any reasonable reconstruction of these bacteria genomes. The anthropological examination of ancient skulls from which the T. -

THE HUMAN MICROBIOTA: the ROLE of MICROBIAL COMMUNITIES in HEALTH and DISEASE La Microbiota Humana: Comunidades Microbianas En La Salud Y En La Enfermedad
ACTA BIOLÓGICA COLOMBIANA http://www.revistas.unal.edu.co/index.php/actabiol SEDE BOGOTÁ FACULTAD DE CIENCIAS ARTÍCULODEPARTAMENTO DE DE REVISIÓN BIOLOGÍA INVITADO / INVITED REVIEW THE HUMAN MICROBIOTA: THE ROLE OF MICROBIAL COMMUNITIES IN HEALTH AND DISEASE La microbiota humana: comunidades microbianas en la salud y en la enfermedad Luz Elena BOTERO1,2, Luisa DELGADO-SERRANO3,4, Martha Lucía CEPEDA HERNÁNDEZ3, Patricia DEL PORTILLO OBANDO3, María Mercedes ZAMBRANO EDER3. 1 Facultad de Medicina, Universidad Pontificia Bolivariana. Calle 78B no. 72A-109, Bloque B, Piso 5. Medellín, Colombia. 2 Unidad de Bacteriología y Micobacterias, Corporación para Investigaciones Biológicas, Unidad Pontificia Bolivariana. Carrera 72A no. 78B-141. Medellín, Colombia. 3 Corporación Corpogen. Carrera 5 no. 66A-34. Bogotá D.C., Colombia. 4 Centro de Bioinformática y Biología Computacional- BIOS. Carrera 15B no. 161. Manizales, Colombia. For correspondence. [email protected] Received: 22nd March 2015, Returned for revision: 14th April 2015, Accepted: 10th July 2015. Associate Editor: Nubia Estela Matta Camacho. Citation / Citar este artículo como: Botero LE, Delgado-Serrano L, Cepeda ML, Del Portillo P, Zambrano MM. The human microbiota: the role of microbial communities in health and disease. Acta biol. Colomb. 2016;21(1):5-15. doi: http://dx.doi.org/10.15446/abc.v21n1.49761 ABSTRACT During the last decade, there has been increasing awareness of the massive number of microorganisms, collectively known as the human microbiota, that are associated with humans. This microbiota outnumbers the host cells by approximately a factor of ten and contains a large repertoire of microbial genome-encoded metabolic processes. The diverse human microbiota and its associated metabolic potential can provide the host with novel functions that can influence host health and disease status in ways that still need to be analyzed. -
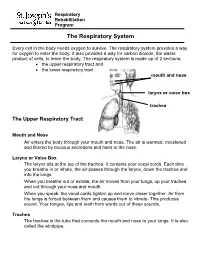
The Respiratory System
Respiratory Rehabilitation Program The Respiratory System Every cell in the body needs oxygen to survive. The respiratory system provides a way for oxygen to enter the body. It also provides a way for carbon dioxide, the waste product of cells, to leave the body. The respiratory system is made up of 2 sections: the upper respiratory tract and the lower respiratory tract mouth and nose larynx or voice box trachea The Upper Respiratory Tract Mouth and Nose Air enters the body through your mouth and nose. The air is warmed, moistened and filtered by mucous secretions and hairs in the nose. Larynx or Voice Box The larynx sits at the top of the trachea. It contains your vocal cords. Each time you breathe in or inhale, the air passes through the larynx, down the trachea and into the lungs. When you breathe out or exhale, the air moves from your lungs, up your trachea and out through your nose and mouth. When you speak, the vocal cords tighten up and move closer together. Air from the lungs is forced between them and causes them to vibrate. This produces sound. Your tongue, lips and teeth form words out of these sounds. Trachea The trachea is the tube that connects the mouth and nose to your lungs. It is also called the windpipe. The Lower Respiratory Tract Inside Lungs Outside Lungs bronchial tubes alveoli diaphragm (muscle) Bronchial Tubes The trachea splits into 2 bronchial tubes in your lungs. These are called the left bronchus and right bronchus. The bronchus tubes keep branching off into smaller and smaller tubes called bronchi. -

From Donor to Patient: Collection, Preparation and Cryopreservation of Fecal Samples for Fecal Microbiota Transplantation
diseases Review From Donor to Patient: Collection, Preparation and Cryopreservation of Fecal Samples for Fecal Microbiota Transplantation Carole Nicco 1 , Armelle Paule 2, Peter Konturek 3 and Marvin Edeas 1,* 1 Cochin Institute, INSERM U1016, University Paris Descartes, Development, Reproduction and Cancer, Cochin Hospital, 75014 Paris, France; [email protected] 2 International Society of Microbiota, 75002 Paris, France; [email protected] 3 Teaching Hospital of the University of Jena, Thuringia-Clinic Saalfeld, 07318 Saalfeld, Germany; [email protected] * Correspondence: [email protected] Received: 6 March 2020; Accepted: 10 April 2020; Published: 15 April 2020 Abstract: Fecal Microbiota Transplantation (FMT) is suggested as an efficacious therapeutic strategy for restoring intestinal microbial balance, and thus for treating disease associated with alteration of gut microbiota. FMT consists of the administration of fresh or frozen fecal microorganisms from a healthy donor into the intestinal tract of diseased patients. At this time, in according to healthcare authorities, FMT is mainly used to treat recurrent Clostridium difficile. Despite the existence of a few existing stool banks worldwide and many studies of the FMT, there is no standard method for producing material for FMT, and there are a multitude of factors that can vary between the institutions. The main constraints for the therapeutic uses of FMT are safety concerns and acceptability. Technical and logistical issues arise when establishing such a non-standardized treatment into clinical practice with safety and proper governance. In this context, our manuscript describes a process of donor safety screening for FMT compiling clinical and biological examinations, questionnaires and interviews of donors.