Jukes-Cantor Evolutionary Model Based Phylogenetic Relationship of Economically Important Ornamental Palms Using Maximum Likelihood Approach - 14859
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Journal of the International Palm Society Vol. 58(4) Dec. 2014 the INTERNATIONAL PALM SOCIETY, INC
Palms Journal of the International Palm Society Vol. 58(4) Dec. 2014 THE INTERNATIONAL PALM SOCIETY, INC. The International Palm Society Palms (formerly PRINCIPES) Journal of The International Palm Society Founder: Dent Smith The International Palm Society is a nonprofit corporation An illustrated, peer-reviewed quarterly devoted to engaged in the study of palms. The society is inter- information about palms and published in March, national in scope with worldwide membership, and the June, September and December by The International formation of regional or local chapters affiliated with the Palm Society Inc., 9300 Sandstone St., Austin, TX international society is encouraged. Please address all 78737-1135 USA. inquiries regarding membership or information about Editors: John Dransfield, Herbarium, Royal Botanic the society to The International Palm Society Inc., 9300 Gardens, Kew, Richmond, Surrey, TW9 3AE, United Sandstone St., Austin, TX 78737-1135 USA, or by e-mail Kingdom, e-mail [email protected], tel. 44-20- to [email protected], fax 512-607-6468. 8332-5225, Fax 44-20-8332-5278. OFFICERS: Scott Zona, Dept. of Biological Sciences (OE 167), Florida International University, 11200 SW 8 Street, President: Leland Lai, 21480 Colina Drive, Topanga, Miami, Florida 33199 USA, e-mail [email protected], tel. California 90290 USA, e-mail [email protected], 1-305-348-1247, Fax 1-305-348-1986. tel. 1-310-383-2607. Associate Editor: Natalie Uhl, 228 Plant Science, Vice-Presidents: Jeff Brusseau, 1030 Heather Drive, Cornell University, Ithaca, New York 14853 USA, e- Vista, California 92084 USA, e-mail mail [email protected], tel. 1-607-257-0885. -

Nymphalidae, Brassolinae) from Panama, with Remarks on Larval Food Plants for the Subfamily
Journal of the Lepidopterists' Society 5,3 (4), 1999, 142- 152 EARLY STAGES OF CALICO ILLIONEUS AND C. lDOMENEUS (NYMPHALIDAE, BRASSOLINAE) FROM PANAMA, WITH REMARKS ON LARVAL FOOD PLANTS FOR THE SUBFAMILY. CARLA M. PENZ Department of Invertebrate Zoology, Milwaukee Public Museum, 800 West Wells Street, Milwaukee, Wisconsin 53233, USA , and Curso de P6s-Gradua9ao em Biocicncias, Pontiffcia Universidade Cat61ica do Rio Grande do SuI, Av. Ipiranga 6681, FOlto Alegre, RS 90619-900, BRAZIL ANNETTE AIELLO Smithsonian Tropical Research Institute, Apdo. 2072, Balboa, Ancon, HEPUBLIC OF PANAMA AND ROBERT B. SRYGLEY Smithsonian Tropical Research Institute, Apdo. 2072, Balboa, Ancon, REPUBLIC OF PANAMA, and Department of Zoology, University of Oxford, South Parks Road, Oxford, OX13PS, ENGLAND ABSTRACT, Here we describe the complete life cycle of Galigo illioneus oberon Butler and the mature larva and pupa of C. idomeneus (L.). The mature larva and pupa of each species are illustrated. We also provide a compilation of host records for members of the Brassolinae and briefly address the interaction between these butterflies and their larval food plants, Additional key words: Central America, host records, monocotyledonous plants, larval food plants. The nymphalid subfamily Brassolinae includes METHODS Neotropical species of large body size and crepuscular habits, both as caterpillars and adults (Harrison 1963, Between 25 May and .31 December, 1994 we Casagrande 1979, DeVries 1987, Slygley 1994). Larvae searched for ovipositing female butterflies along generally consume large quantities of plant material to Pipeline Road, Soberania National Park, Panama, mo reach maturity, a behavior that may be related as much tivated by a study on Caligo mating behavior (Srygley to the low nutrient content of their larval food plants & Penz 1999). -

Coevolution of Cycads and Dinosaurs George E
Coevolution of cycads and dinosaurs George E. Mustoe* INTRODUCTION TOXICOLOGY OF EXTANT CYCADS cycads suggests that the biosynthesis of ycads were a major component of Illustrations in textbooks commonly these compounds was a trait that C forests during the Mesozoic Era, the depict herbivorous dinosaurs browsing evolved early in the history of the shade of their fronds falling upon the on cycad fronds, but biochemical evi- Cycadales. Brenner et al. (2002) sug- scaly backs of multitudes of dinosaurs dence from extant cycads suggests that gested that macrozamin possibly serves a that roamed the land. Paleontologists these reconstructions are incorrect. regulatory function during cycad have long postulated that cycad foliage Foliage of modern cycads is highly toxic growth, but a strong case can be made provided an important food source for to vertebrates because of the presence that the most important reason for the reptilian herbivores, but the extinction of two powerful neurotoxins and carcin- evolution of cycad toxins was their of dinosaurs and the contemporaneous ogens, cycasin (methylazoxymethanol- usefulness as a defense against foliage precipitous decline in cycad popula- beta-D-glucoside) and macrozamin (beta- predation at a time when dinosaurs were tions at the close of the Cretaceous N-methylamine-L-alanine). Acute symp- the dominant herbivores. The protective have generally been assumed to have toms triggered by cycad foliage inges- role of these toxins is evidenced by the resulted from different causes. Ecologic tion include vomiting, diarrhea, and seed dispersal characteristics of effects triggered by a cosmic impact are abdominal cramps, followed later by loss modern cycads. a widely-accepted explanation for dino- of coordination and paralysis of the saur extinction; cycads are presumed to limbs. -

Rhyzobius Lophanthae Introduced Against Asian
ABSTRACT Too Little and Too Late???? Asian Cycad Scale (ACS) Chronology Asian cycad scale (ACS), Aulacaspis yasumatsui, was 1972 – Aulacaspis yasumatsui described in Thailand first detected in Tumon, Guam in December 2003 in front Rhyzobius lophanthae introduced against Asian 1996 – ACS detected in Florida of a hotel where Cycas revoluta, an introduced ornamental 1998 – ACS detected in Hawaii cycad and Cycas micronesica, an indigenous cycad were cycad scale, Aulacaspis yasumatsui, on Guam 2003 – ACS detected on cycads used for landscaping in Guam’s planted. The scale is believed to have been imported from Tumon Bay hotel district Hawaii in 1998 on ornamental cycads. The scale currently R.H. Miller1, A. Moore1, R.N. Muniappan1, A.P. Brooke2 and T.E. Marler1. 2004 – ACS spreads to Cycas revoluta and C. micronesica infests introduced and indigenous cycads on about two 1CNAS-AES, University of Guam, Mangilao, Guam (fadang) throughout Guam thirds of Guam’s 354 square kilometers. Severe 2Guam National Wildlife Refuge, Dededo, Guam 2005 – Ryzobius lophanthae and Coccobius fulvus released on infestations have been observed to kill both species within Guam; Plans made to preserve C. micronesica germplasm from a few months. We fear that C. micronesica may be Guam on the nearby island of Tinian threatened with extinction should the scale spread to the few other Micronesian islands that harbor it. Rhyzobius lophanthae, a coccinellid introduced to Asian Cycad Scale Management Hawaii in 1894 for other scale insects, was imported from Biological Control Agents on Guam Maui to Guam in November 2004 and released on C. Rhyzobius lophanthae micronesica at the Guam National Wildlife Refuge at • Introduced in Hawaii in 1894; Guam ??? 1930s Ritidian point in February 2005. -
Red Palm Mite)
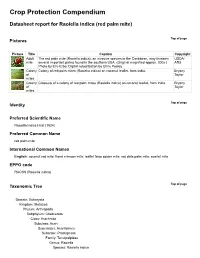
Crop Protection Compendium Datasheet report for Raoiella indica (red palm mite) Top of page Pictures Picture Title Caption Copyright Adult The red palm mite (Raoiella indica), an invasive species in the Caribbean, may threaten USDA- mite several important palms found in the southern USA. (Original magnified approx. 300x.) ARS Photo by Eric Erbe; Digital colourization by Chris Pooley. Colony Colony of red palm mites (Raoiella indica) on coconut leaflet, from India. Bryony of Taylor mites Colony Close-up of a colony of red palm mites (Raoiella indica) on coconut leaflet, from India. Bryony of Taylor mites Top of page Identity Preferred Scientific Name Raoiella indica Hirst (1924) Preferred Common Name red palm mite International Common Names English: coconut red mite; frond crimson mite; leaflet false spider mite; red date palm mite; scarlet mite EPPO code RAOIIN (Raoiella indica) Top of page Taxonomic Tree Domain: Eukaryota Kingdom: Metazoa Phylum: Arthropoda Subphylum: Chelicerata Class: Arachnida Subclass: Acari Superorder: Acariformes Suborder: Prostigmata Family: Tenuipalpidae Genus: Raoiella Species: Raoiella indica / Top of page Notes on Taxonomy and Nomenclature R. indica was first described in the district of Coimbatore (India) by Hirst in 1924 on coconut leaflets [Cocos nucifera]. A comprehensive taxonomic review of the genus and species was carried out by Mesa et al. (2009), which lists all suspected junior synonyms of R. indica, including Raoiella camur (Chaudhri and Akbar), Raoiella empedos (Chaudhri and Akbar), Raoiella obelias (Hasan and Akbar), Raoiella pandanae (Mohanasundaram), Raoiella phoenica (Meyer) and Raoiella rahii (Akbar and Chaudhri). The review also highlighted synonymy with Rarosiella cocosae found on coconut in the Philippines. -

Seed Geometry in the Arecaceae
horticulturae Review Seed Geometry in the Arecaceae Diego Gutiérrez del Pozo 1, José Javier Martín-Gómez 2 , Ángel Tocino 3 and Emilio Cervantes 2,* 1 Departamento de Conservación y Manejo de Vida Silvestre (CYMVIS), Universidad Estatal Amazónica (UEA), Carretera Tena a Puyo Km. 44, Napo EC-150950, Ecuador; [email protected] 2 IRNASA-CSIC, Cordel de Merinas 40, E-37008 Salamanca, Spain; [email protected] 3 Departamento de Matemáticas, Facultad de Ciencias, Universidad de Salamanca, Plaza de la Merced 1–4, 37008 Salamanca, Spain; [email protected] * Correspondence: [email protected]; Tel.: +34-923219606 Received: 31 August 2020; Accepted: 2 October 2020; Published: 7 October 2020 Abstract: Fruit and seed shape are important characteristics in taxonomy providing information on ecological, nutritional, and developmental aspects, but their application requires quantification. We propose a method for seed shape quantification based on the comparison of the bi-dimensional images of the seeds with geometric figures. J index is the percent of similarity of a seed image with a figure taken as a model. Models in shape quantification include geometrical figures (circle, ellipse, oval ::: ) and their derivatives, as well as other figures obtained as geometric representations of algebraic equations. The analysis is based on three sources: Published work, images available on the Internet, and seeds collected or stored in our collections. Some of the models here described are applied for the first time in seed morphology, like the superellipses, a group of bidimensional figures that represent well seed shape in species of the Calamoideae and Phoenix canariensis Hort. ex Chabaud. -

WRA Species Report
Family: Arecaceae Taxon: Caryota urens Synonym: NA Common Name: Fishtail palm Jaggery palm Toddy palm Wine Palm Questionaire : current 20090513 Assessor: Chuck Chimera Designation: EVALUATE Status: Assessor Approved Data Entry Person: Assessor WRA Score 5 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? y=1, n=-1 103 Does the species have weedy races? y=1, n=-1 201 Species suited to tropical or subtropical climate(s) - If island is primarily wet habitat, then (0-low; 1-intermediate; 2- High substitute "wet tropical" for "tropical or subtropical" high) (See Appendix 2) 202 Quality of climate match data (0-low; 1-intermediate; 2- High high) (See Appendix 2) 203 Broad climate suitability (environmental versatility) y=1, n=0 n 204 Native or naturalized in regions with tropical or subtropical climates y=1, n=0 y 205 Does the species have a history of repeated introductions outside its natural range? y=-2, ?=-1, n=0 y 301 Naturalized beyond native range y = 1*multiplier (see y Appendix 2), n= question 205 302 Garden/amenity/disturbance weed n=0, y = 1*multiplier (see y Appendix 2) 303 Agricultural/forestry/horticultural weed n=0, y = 2*multiplier (see n Appendix 2) 304 Environmental weed n=0, y = 2*multiplier (see Appendix 2) 305 Congeneric weed n=0, y = 1*multiplier (see y Appendix 2) 401 Produces spines, thorns or burrs y=1, n=0 n 402 Allelopathic y=1, n=0 403 Parasitic y=1, n=0 n 404 Unpalatable to grazing animals y=1, n=-1 n 405 Toxic to animals y=1, n=0 n 406 Host for recognized pests -

Technical Background Document in Support of the Mid-Term Review of the Global Strategy for Plant Conservation (GSPC)
Technical background document for the mid-term review of the GSPC Technical background document in support of the mid-term review of the Global Strategy for Plant Conservation (GSPC) Compiled by Botanic Gardens Conservation International (BGCI) in association with the Global Partnership for Plant Conservation (GPPC) and the Secretariat of the Convention on Biological Diversity 1 Technical background document for the mid-term review of the GSPC Contents Introduction ......................................................................................................................................5 Section 1: Progress in national / regional implementation of the GSPC ................................................6 The GSPC and National / Regional Biodiversity Strategies and Action Plans ........................................... 6 Progress in plant conservation as reported in 5th National Reports to the CBD ...................................... 7 Reviews from regional workshops ............................................................................................................ 8 Progress in China ....................................................................................................................................... 8 Progress in Brazil ....................................................................................................................................... 9 Progress in Europe ................................................................................................................................. -

1 Ornamental Palms
1 Ornamental Palms: Biology and Horticulture T.K. Broschat and M.L. Elliott Fort Lauderdale Research and Education Center University of Florida, Davie, FL 33314, USA D.R. Hodel University of California Cooperative Extension Alhambra, CA 91801, USA ABSTRACT Ornamental palms are important components of tropical, subtropical, and even warm temperate climate landscapes. In colder climates, they are important interiorscape plants and are often a focal point in malls, businesses, and other public areas. As arborescent monocots, palms have a unique morphology and this greatly influences their cultural requirements. Ornamental palms are over- whelmingly seed propagated, with seeds of most species germinating slowly and being intolerant of prolonged storage or cold temperatures. They generally do not have dormancy requirements, but do require high temperatures (30–35°C) for optimum germination. Palms are usually grown in containers prior to trans- planting into a field nursery or landscape. Because of their adventitious root system, large field-grown specimen palms can easily be transplanted. In the landscape, palm health and quality are greatly affected by nutritional deficien- cies, which can reduce their aesthetic value, growth rate, or even cause death. Palm life canCOPYRIGHTED also be shortened by a number of MATERIAL diseases or insect pests, some of which are lethal, have no controls, or have wide host ranges. With the increasing use of palms in the landscape, pathogens and insect pests have moved with the Horticultural Reviews, Volume 42, First Edition. Edited by Jules Janick. 2014 Wiley-Blackwell. Published 2014 by John Wiley & Sons, Inc. 1 2 T.K. BROSCHAT, D.R. HODEL, AND M.L. -

North American Botanic Garden Strategy for Plant Conservation 2016-2020 Botanic Gardens Conservation International
Nova Southeastern University NSUWorks Marine & Environmental Sciences Faculty Reports Department of Marine and Environmental Sciences 1-1-2016 North American Botanic Garden Strategy for Plant Conservation 2016-2020 Botanic Gardens Conservation International American Public Gardens Association Asociacion Mexicana de Jardines Center for Plant Conservation Plant Conservation Alliance See next page for additional authors Find out more information about Nova Southeastern University and the Halmos College of Natural Sciences and Oceanography. Follow this and additional works at: https://nsuworks.nova.edu/occ_facreports Part of the Plant Sciences Commons Authors Botanic Gardens Conservation International, American Public Gardens Association, Asociacion Mexicana de Jardines, Center for Plant Conservation, Plant Conservation Alliance, Pam Allenstein, Robert Bye, Jennifer Ceska, John Clark, Jenny Cruse-Sanders, Gerard Donnelly, Christopher Dunn, Anne Frances, David Galbraith, Jordan Golubov, Gennadyi Gurman, Kayri Havens, Abby Hird Meyer, Douglas Justice, Edelmira Linares, Maria Magdalena Hernandez, Beatriz Maruri Aguilar, Mike Maunder, Ray Mims, Greg Mueller, Jennifer Ramp Neale, Martin Nicholson, Ari Novy, Susan Pell, John J. Pipoly III, Diane Ragone, Peter Raven, Erin Riggs, Kate Sackman, Emiliano Sanchez Martinez, Suzanne Sharrock, Casey Sclar, Paul Smith, Murphy Westwood, Rebecca Wolf, and Peter Wyse Jackson North American Botanic Garden Strategy For Plant Conservation 2016-2020 North American Botanic Garden Strategy For Plant Conservation 2016-2020 Acknowledgements Published January 2016 by Botanic Gardens Conservation International. Support from the United States Botanic Garden, the American Public Gardens Association, and the Center for Plant Conservation helped make this publication possible. The 2016-2020 North American Botanic Garden Strategy for Plant Conservation is dedicated to the late Steven E. Clemants, who so diligently and ably led the creation of the original North American Strategy published in 2006. -

(Arecaceae): Évolution Du Système Sexuel Et Du Nombre D'étamines
Etude de l’appareil reproducteur des palmiers (Arecaceae) : évolution du système sexuel et du nombre d’étamines Elodie Alapetite To cite this version: Elodie Alapetite. Etude de l’appareil reproducteur des palmiers (Arecaceae) : évolution du système sexuel et du nombre d’étamines. Sciences agricoles. Université Paris Sud - Paris XI, 2013. Français. NNT : 2013PA112063. tel-01017166 HAL Id: tel-01017166 https://tel.archives-ouvertes.fr/tel-01017166 Submitted on 2 Jul 2014 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITE PARIS-SUD ÉCOLE DOCTORALE : Sciences du Végétal (ED 45) Laboratoire d'Ecologie, Systématique et E,olution (ESE) DISCIPLINE : -iologie THÈSE DE DOCTORAT SUR TRAVAUX soutenue le ./05/10 2 par Elodie ALAPETITE ETUDE DE L'APPAREIL REPRODUCTEUR DES PAL4IERS (ARECACEAE) : EVOLUTION DU S5STE4E SE6UEL ET DU NO4-RE D'ETA4INES Directeur de thèse : Sophie NADOT Professeur (Uni,ersité Paris-Sud Orsay) Com osition du jury : Rapporteurs : 9ean-5,es DU-UISSON Professeur (Uni,ersité Pierre et 4arie Curie : Paris VI) Porter P. LOWR5 Professeur (4issouri -otanical Garden USA et 4uséum National d'Histoire Naturelle Paris) Examinateurs : Anders S. -ARFOD Professeur (Aarhus Uni,ersity Danemark) Isabelle DA9OA Professeur (Uni,ersité Paris Diderot : Paris VII) 4ichel DRON Professeur (Uni,ersité Paris-Sud Orsay) 3 4 Résumé Les palmiers constituent une famille emblématique de monocotylédones, comprenant 183 genres et environ 2500 espèces distribuées sur tous les continents dans les zones tropicales et subtropicales. -

Zamiaceae, Cycadales) and Evolution in Cycadales
The complete chloroplast genome of Microcycas calocoma (Miq.) A. DC. (Zamiaceae, Cycadales) and evolution in Cycadales Aimee Caye G. Chang1,2,3, Qiang Lai1, Tao Chen2, Tieyao Tu1, Yunhua Wang2, Esperanza Maribel G. Agoo4, Jun Duan1 and Nan Li2 1 South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China 2 Shenzhen Fairy Lake Botanical Garden, Chinese Academy of Sciences, Shenzhen, China 3 University of Chinese Academy of Sciences, Beijing, China 4 Department of Biology, De La Salle University, Manila, Philippines ABSTRACT Cycadales is an extant group of seed plants occurring in subtropical and tropical regions comprising putatively three families and 10 genera. At least one complete plastid genome sequence has been reported for all of the 10 genera except Microcycas, making it an ideal plant group to conduct comprehensive plastome comparisons at the genus level. This article reports for the first time the plastid genome of Microcycas calocoma. The plastid genome has a length of 165,688 bp with 134 annotated genes including 86 protein-coding genes, 47 non-coding RNA genes (39 tRNA and eight rRNA) and one pseudogene. Using global sequence variation analysis, the results showed that all cycad genomes share highly similar genomic profiles indicating significant slow evolution and little variation. However, identity matrices coinciding with the inverted repeat regions showed fewer similarities indicating that higher polymorphic events occur at those sites. Conserved non-coding regions also appear to be more divergent whereas variations in the exons were less discernible indicating that the latter comprises more conserved sequences. Submitted 5 September 2019 Phylogenetic analysis using 81 concatenated protein-coding genes of chloroplast (cp) Accepted 27 November 2019 genomes, obtained using maximum likelihood and Bayesian inference with high Published 13 January 2020 support values (>70% ML and = 1.0 BPP), confirms that Microcycas is closest to Corresponding authors Zamia and forms a monophyletic clade with Ceratozamia and Stangeria.