Is Generation of C3(H2O) Necessary for Activation of the Alternative Pathway T in Real Life? ⁎ Kristina N
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Haemostasis Antibodies from Cedarlane
HAEMOSTASIS ANTIBODIES FROM CEDARLANE Haemostasis Antibodies www.cedarlanelabs.com Haemostasis Antibodies The study of haemostasis and thrombosis has become one of the most rapidly growing fields in medical research. An unprecedented amount of research is currently focused on the study of the regulatory mechanisms involved in blood clot formation, subsequent lysis, and potential pharmacological intervention of these processes. Cedarlane offers a complete line of antibodies to haemostasis related antigens. The high quality and purity of these antibodies make them valuable tools for applications including ELISA, immunohistochemistry, immunoblotting (Western blot) and immunoprecipitation techniques (including immunodiffusion and rocket immunoelectrophoresis). Haemostasis Antibodies Haemostasis NEW! Cedarlane offers an expanding range of new monoclonal antibodies to various haemostasis related antigens, providing customers a more comprehensive array of tools for the study of haemostasis and thrombosis. These antibodies have been tested for suitability for use in Western Blotting and ELISA. Featured Antibodies Specificity Format Clone Isotype Species Application Size Cat # Price $ Reactivity Factor VII/VIIA Purified AA-3 mouse IgG1 Human E, WB 200 μg CL2763AP 259 Factor VII/VIIA Biotin AA-3 mouse IgG1 Human E, WB 100 μg CL2763B 199 Factor VII/VIIa Purified AD-1 mouse IgG1 Human E, WB 200 μg CL2764AP 259 Factor VII/VIIa Biotin AD-1 mouse IgG1 Human E, WB 100 μg CL2764B 199 Factor VII/VIIa (Calcium Dependent) Purified CaFVII-22 mouse IgG1 Human -

Plasma Contact System Activation Drives Anaphylaxis in Severe Mast Cell–Mediated Allergic Reactions
Plasma contact system activation drives anaphylaxis in severe mast cell–mediated allergic reactions Anna Sala-Cunill, MD, PhD,a,b,c Jenny Bjorkqvist,€ MSc,c,d Riccardo Senter, MD,c,e Mar Guilarte, MD, PhD,a,b Victoria Cardona, MD, PhD,a,b Moises Labrador, MD, PhD,a,b Katrin F. Nickel, PhD,c,d,f Lynn Butler, PhD,c,d,f Olga Luengo, MD, PhD,a,b Parvin Kumar, MSc,c,d Linda Labberton, MSc,c,d Andy Long, PhD,f Antonio Di Gennaro, PhD,c,d Ellinor Kenne, PhD,c,d Anne Jams€ a,€ PhD,c,d Thorsten Krieger, MD,f Hartmut Schluter,€ PhD,f Tobias Fuchs, PhD,c,d,f Stefanie Flohr, PhD,g Ulrich Hassiepen, PhD,g Frederic Cumin, PhD,g Keith McCrae, MD,h Coen Maas, PhD,i Evi Stavrou, MD,j and Thomas Renne, MD, PhDc,d,f Barcelona, Spain, Stockholm, Sweden, Padua, Italy, Hamburg, Germany, Basel, Switzerland, Cleveland, Ohio, and Utrecht, The Netherlands Background: Anaphylaxis is an acute, potentially lethal, hypotension. Activated mast cells systemically released heparin, multisystem syndrome resulting from the sudden release of mast which provided a negatively charged surface for factor XII cell–derived mediators into the circulation. autoactivation. Activated factor XII generates plasma Objectives and Methods: We report here that a plasma protease kallikrein, which proteolyzes kininogen, leading to the cascade, the factor XII–driven contact system, critically liberation of bradykinin. We evaluated the contact system in contributes to the pathogenesis of anaphylaxis in both murine patients with anaphylaxis. In all 10 plasma samples models and human subjects. immunoblotting revealed activation of factor XII, plasma Results: Deficiency in or pharmacologic inhibition of factor XII, kallikrein, and kininogen during the acute phase of anaphylaxis plasma kallikrein, high-molecular-weight kininogen, or the but not at basal conditions or in healthy control subjects. -

SARS-Cov-2 Entry Protein TMPRSS2 and Its Homologue, TMPRSS4
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441280; this version posted April 26, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 SARS-CoV-2 Entry Protein TMPRSS2 and Its 2 Homologue, TMPRSS4 Adopts Structural Fold Similar 3 to Blood Coagulation and Complement Pathway 4 Related Proteins ∗,a ∗∗,b b 5 Vijaykumar Yogesh Muley , Amit Singh , Karl Gruber , Alfredo ∗,a 6 Varela-Echavarría a 7 Instituto de Neurobiología, Universidad Nacional Autónoma de México, Querétaro, México b 8 Institute of Molecular Biosciences, University of Graz, Graz, Austria 9 Abstract The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) utilizes TMPRSS2 receptor to enter target human cells and subsequently causes coron- avirus disease 19 (COVID-19). TMPRSS2 belongs to the type II serine proteases of subfamily TMPRSS, which is characterized by the presence of the serine- protease domain. TMPRSS4 is another TMPRSS member, which has a domain architecture similar to TMPRSS2. TMPRSS2 and TMPRSS4 have been shown to be involved in SARS-CoV-2 infection. However, their normal physiological roles have not been explored in detail. In this study, we analyzed the amino acid sequences and predicted 3D structures of TMPRSS2 and TMPRSS4 to under- stand their functional aspects at the protein domain level. Our results suggest that these proteins are likely to have common functions based on their conserved domain organization. -

Identification of New Substrates and Physiological Relevance
Université de Montréal The Multifaceted Proprotein Convertases PC7 and Furin: Identification of New Substrates and Physiological Relevance Par Stéphanie Duval Biologie Moléculaire, Faculté de médecine Thèse présentée en vue de l’obtention du grade de Philosophiae doctor (Ph.D) en Biologie moléculaire, option médecine cellulaire et moléculaire Avril 2020 © Stéphanie Duval, 2020 Résumé Les proprotéines convertases (PCs) sont responsables de la maturation de plusieurs protéines précurseurs et sont impliquées dans divers processus biologiques importants. Durant les 30 dernières années, plusieurs études sur les PCs se sont traduites en succès cliniques, toutefois les fonctions spécifiques de PC7 demeurent obscures. Afin de comprendre PC7 et d’identifier de nouveaux substrats, nous avons généré une analyse protéomique des protéines sécrétées dans les cellules HuH7. Cette analyse nous a permis d’identifier deux protéines transmembranaires de fonctions inconnues: CASC4 et GPP130/GOLIM4. Au cours de cette thèse, nous nous sommes aussi intéressé au rôle de PC7 dans les troubles comportementaux, grâce à un substrat connu, BDNF. Dans le chapitre premier, je présenterai une revue de la littérature portant entre autres sur les PCs. Dans le chapitre II, l’étude de CASC4 nous a permis de démontrer que cette protéine est clivée au site KR66↓NS par PC7 et Furin dans des compartiments cellulaires acides. Comme CASC4 a été rapporté dans des études de cancer du sein, nous avons généré des cellules MDA- MB-231 exprimant CASC4 de type sauvage et avons démontré une diminution significative de la migration et de l’invasion cellulaire. Ce phénotype est causé notamment par une augmentation du nombre de complexes d’adhésion focale et peut être contrecarré par la surexpression d’une protéine CASC4 mutante ayant un site de clivage optimale par PC7/Furin ou encore en exprimant une protéine contenant uniquement le domaine clivé N-terminal. -

An Overview of the Kallikrein Gene Families in Humans and Other Species: Emerging Candidate Tumour Markers૾
Clinical Biochemistry 36 (2003) 443–452 An overview of the kallikrein gene families in humans and other species: Emerging candidate tumour markers૾ George M. Yousefa,b, Eleftherios P. Diamandisa,b,* aDepartment of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario, Canada bDepartment of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario, Canada Abstract Kallikreins are serine proteases with diverse physiologic functions. They are represented by multigene families in many animal species, especially in rat and mouse. Recently, the human kallikrein gene family has been fully characterized and includes 15 members, tandemly localized on chromosome 19q13.4. A new definition has now been proposed for kallikreins, which is not based on function but, rather, on close proximity and structural similarities. In this review, we summarize available information about kallikreins in many animal species with special emphasis on human kallikreins. We discuss the common structural features of kallikreins at the DNA, mRNA and protein levels and overview their evolutionary history. Kallikreins are expressed in a wide range of tissues including the salivary gland, endocrine or endocrine-related tissues such as testis, prostate, breast and endometrium and in the central nervous system. Most, if not all, genes are under steroid hormone regulation. Accumulating evidence indicates that kallikreins are involved in many pathologic conditions. Of special interest is the potential role of kallikreins in the central nervous system. In addition, many kallikreins seem to be candidate tumor markers for many malignancies, especially those of endocrine-related organs. © 2003 The Canadian Society of Clinical Chemists. All rights reserved. Keywords: Kallikrein; Tumor markers; Cancer biomarkers; Prostate cancer; Breast cancer; Ovarian cancer; Alzheimer’s disease; Serine proteases; Chromosome 19; Kallikrein evolution; Rodent kallikreins; Hormonally regulated genes 1. -

Downloaded from Genomic Data Common Website (GDC at Accessed on 2019)
G C A T T A C G G C A T genes Article Molecular Pathways Associated with Kallikrein 6 Overexpression in Colorectal Cancer Ritu Pandey 1,2,*, Muhan Zhou 3, Yuliang Chen 3, Dalila Darmoul 4 , Conner C. Kisiel 2, Valentine N. Nfonsam 5 and Natalia A. Ignatenko 1,2 1 Department of Cellular and Molecular Medicine, University of Arizona, Tucson, AZ 85721, USA; [email protected] 2 University of Arizona Cancer Center, University of Arizona, Tucson, AZ 85724, USA; [email protected] 3 Bioinformatics Shared Resource, University of Arizona Cancer Center, Tucson, AZ 85724, USA; [email protected] (M.Z.); [email protected] (Y.C.) 4 Institut National de la Santé et de la Recherche Médicale (INSERM), Université de Paris, Lariboisière Hospital, 75010 Paris, France; [email protected] 5 Department of Surgery, Section of Surgical Oncology, University of Arizona, Tucson, AZ 85724, USA; [email protected] * Correspondence: [email protected] Abstract: Colorectal cancer (CRC) remains one of the leading causes of cancer-related death world- wide. The high mortality of CRC is related to its ability to metastasize to distant organs. The kallikrein-related peptidase Kallikrein 6 (KLK6) is overexpressed in CRC and contributes to cancer cell invasion and metastasis. The goal of this study was to identify KLK6-associated markers for the CRC prognosis and treatment. Tumor Samples from the CRC patients with significantly elevated Citation: Pandey, R.; Zhou, M.; Chen, KLK6 transcript levels were identified in the RNA-Seq data from Cancer Genome Atlas (TCGA) Y.; Darmoul, D.; Kisiel, C.C.; and their expression profiles were evaluated using Gene Ontology (GO), Phenotype and Reactome Nfonsam, V.N.; Ignatenko, N.A. -

Cloning of a New Member of the Human Kallikrein Gene Family, KLK14, Which Is Down-Regulated in Different Malignancies
[CANCER RESEARCH 61, 3425–3431, April 15, 2001] Cloning of a New Member of the Human Kallikrein Gene Family, KLK14, Which Is Down-Regulated in Different Malignancies George M. Yousef, Angeliki Magklara, Albert Chang, Klaus Jung, Dionyssios Katsaros, and Eleftherios P. Diamandis1 Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada [G. M. Y., A. M., A. C., E. P. D.]; Department of Laboratory Medicine and Pathobiology, University of Toronto, Ontario M5G 1LS, Canada [G. M. Y., A. M., A. C., E. P. D.]; Department of Urology, University Hospital Charite, Humboldt University, Berlin D-10098, Germany [K. J.]; and Department of Gynecologic Oncology, Institute of Obstetrics and Gynecology, University of Turin, Turin 10128, Italy [D. K.] ABSTRACT KLK gene locus and cloned a new gene, KLK14. Here, we describe the cloning of the new gene, its genomic and mRNA structure, its Kallikreins (KLKs) belong to the serine protease family of proteolytic precise location in relation to other known KLKs, and its tissue enzymes. Human pancreatic/renal KLK (KLK1) encodes for an enzyme expression pattern. KLK14 is highly expressed in tissues of the CNS that is involved in posttranslational processing of polypeptide precursors. The function of the other members of this gene family is currently including the brain, cerebellum, and spinal cord. Our preliminary unknown, but growing evidence suggests that many KLKs are implicated results suggest that this gene is down-regulated, at the mRNA level, in in carcinogenesis. By using the positional candidate approach, we were breast, testicular, ovarian, and prostate cancer. able to identify a new human KLK-like gene, KLK14 (also known as KLK-L6). -

Kallikrein 13: a New Player in Coronaviral Infections
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.01.971499; this version posted March 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Kallikrein 13: a new player in coronaviral infections. 2 3 Aleksandra Milewska1,2, Katherine Falkowski2, Magdalena Kalinska3, Ewa Bielecka3, 4 Antonina Naskalska1, Pawel Mak4, Adam Lesner5, Marek Ochman6, Maciej Urlik6, Jan 5 Potempa2,7, Tomasz Kantyka3,8, Krzysztof Pyrc1,* 6 7 1 Virogenetics Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian 8 University, Gronostajowa 7a, 30-387 Krakow, Poland. 9 2 Microbiology Department, Faculty of Biochemistry, Biophysics and Biotechnology, 10 Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland. 11 3 Laboratory of Proteolysis and Post-translational Modification of Proteins, Malopolska 12 Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, 13 Poland. 14 4 Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and 15 Biotechnology, Jagiellonian University, Gronostajowa 7 St., 30-387, Krakow, Poland. 16 5 University of Gdansk, Faculty of Chemistry, Wita Stwosza 63, 80-308 Gdansk, Poland. 17 6 Department of Cardiac, Vascular and Endovascular Surgery and Transplantology, Medical 18 University of Silesia in Katowice, Silesian Centre for Heart Diseases, Zabrze, Poland. 19 7 Centre for Oral Health and Systemic Diseases, University of Louisville School of Dentistry, 20 Louisville, KY 40202, USA. 21 8 Broegelmann Research Laboratory, Department of Clinical Science, University of Bergen, 22 5020 Bergen, Norway 23 24 25 26 27 28 29 30 31 * Correspondence should be addressed to Krzysztof Pyrc ([email protected]), Virogenetics 32 Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian University, 33 Gronostajowa 7, 30-387 Krakow, Poland; Phone number: +48 12 664 61 21; www: 34 http://virogenetics.info/. -
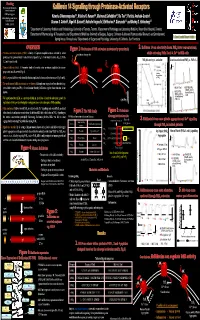
AACR2006-1686P
Funding Proteinases and Inflammation Network Group grant Kallikrein 14 Signalling through Proteinase-Activated Receptors CIHR operating grant 1,2 3,4 3,4 3,4 5 Alberta Heritage Foundation for Katerina Oikonomopoulou , Kristina K. Hansen , Mahmoud Saifeddine , Illa Tea , Patricia Andrade-Gordon , Medical Research Graeme S. Cottrell6, Nigel W. Bunnett6, Nathalie Vergnolle3, Eleftherios P. Diamandis1,2 and Morley D. Hollenberg3,4 NSERC / CRSNG 1Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto; 2Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto; 3Department of Pharmacology & Therapeutics, and 4Department of Medicine, University of Calgary, Calgary; 5Johnson & Johnson Pharmaceutical Research and Development, Spring House, Pennsylvania, 6Departments of Surgery and Physiology, University of California, San Francisco Kallikrein 14 can selectively disarm PAR (lower concentrations), OVERVIEW Figure 1: Mechanism of PAR activation (activation by proteolysis) 2. Kallikrein 14 can selectively disarm PAR1 (lower concentrations), 2+ Proteinase-activated receptors (PARs): a family of G-protein coupled receptors activated by serine proteinase cleavage site whilst activating PARs 2 and 4; Ca in HEK cells proteinases via a proteolytically revealed ‘tethered ligand’ (Fig. 1). Four family members (Fig. 2); PARs N PAR disarming 1, 2 and 4 signal to cells. N PAR1 dis-arming1 vs. activation Selective activation of PAR4 vs. PARs 1, 2 Human kallikreins (hKs): A 15-member family of secreted serine proteinases implicated in tumour 1 cm Thrombin 1 cm 2 min 2 min TFLLR-NH2 progression and cell survival (Fig. 4). (selective desensitization hK14: a tryptic kallikrein; wide tissue distribution, implicated in breast and ovarian cancer (Fig. 5 and 6). of PAR1) The mechanism of kallikrein action is not yet known: Although some targets have been identified (e.g. -

Solenodon Genome Reveals Convergent Evolution of Venom in Eulipotyphlan Mammals
Solenodon genome reveals convergent evolution of venom in eulipotyphlan mammals Nicholas R. Casewella,1, Daniel Petrasb,c, Daren C. Cardd,e,f, Vivek Suranseg, Alexis M. Mychajliwh,i,j, David Richardsk,l, Ivan Koludarovm, Laura-Oana Albulescua, Julien Slagboomn, Benjamin-Florian Hempelb, Neville M. Ngumk, Rosalind J. Kennerleyo, Jorge L. Broccap, Gareth Whiteleya, Robert A. Harrisona, Fiona M. S. Boltona, Jordan Debonoq, Freek J. Vonkr, Jessica Alföldis, Jeremy Johnsons, Elinor K. Karlssons,t, Kerstin Lindblad-Tohs,u, Ian R. Mellork, Roderich D. Süssmuthb, Bryan G. Fryq, Sanjaya Kuruppuv,w, Wayne C. Hodgsonv, Jeroen Kooln, Todd A. Castoed, Ian Barnesx, Kartik Sunagarg, Eivind A. B. Undheimy,z,aa, and Samuel T. Turveybb aCentre for Snakebite Research & Interventions, Liverpool School of Tropical Medicine, Pembroke Place, L3 5QA Liverpool, United Kingdom; bInstitut für Chemie, Technische Universität Berlin, 10623 Berlin, Germany; cCollaborative Mass Spectrometry Innovation Center, University of California, San Diego, La Jolla, CA 92093; dDepartment of Biology, University of Texas at Arlington, Arlington, TX 76010; eDepartment of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138; fMuseum of Comparative Zoology, Harvard University, Cambridge, MA 02138; gEvolutionary Venomics Lab, Centre for Ecological Sciences, Indian Institute of Science, 560012 Bangalore, India; hDepartment of Biology, Stanford University, Stanford, CA 94305; iDepartment of Rancho La Brea, Natural History Museum of Los Angeles County, Los Angeles, -

Aberrant Human Tissue Kallikrein Levels in the Stratum Corneum and Serum of Patients with Psoriasis: Dependence on Phenotype, Severity and Therapy N
CLINICAL AND LABORATORY INVESTIGATIONS DOI 10.1111/j.1365-2133.2006.07743.x Aberrant human tissue kallikrein levels in the stratum corneum and serum of patients with psoriasis: dependence on phenotype, severity and therapy N. Komatsu,* à K. Saijoh,§ C. Kuk,* F. Shirasaki,à K. Takeharaà and E.P. Diamandis* *Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario M5G 1L5, Canada àDepartment of Dermatology and §Department of Hygiene, Graduate School of Medical Science, School of Medicine, Kanazawa University, Kanazawa, Japan Summary Correspondence Background Human tissue kallikreins (KLKs) are a family of 15 trypsin-like or Eleftherios P. Diamandis. chymotrypsin-like secreted serine proteases (KLK1–KLK15). Multiple KLKs have E-mail: [email protected] been quantitatively identified in normal stratum corneum (SC) and sweat as can- didate desquamation-related proteases. Accepted for publication 7 November 2006 Objectives To quantify KLK5, KLK6, KLK7, KLK8, KLK10, KLK11, KLK13 and KLK14 in the SC and serum of patients with psoriasis, and their variation Key words between lesional and nonlesional areas and with phenotype, therapy and severity. diagnostic marker, human kallikreins, psoriasis, The overall SC serine protease activities were also measured. serine proteases, stratum corneum, therapy Methods Enzyme-linked immunosorbent assays and enzymatic assays were used. Conflicts of interest Results The lesional SC of psoriasis generally contained significantly higher levels None declared. of all KLKs. KLK6, KLK10 and KLK13 levels were significantly elevated even in the nonlesional SC. The overall trypsin-like, plasmin-like and furin-like activities were significantly elevated in the lesional SC. -

Salivary Protein Panel to Diagnose Systolic Heart Failure
biomolecules Article Salivary Protein Panel to Diagnose Systolic Heart Failure Xi Zhang 1 , Daniel Broszczak 2, Karam Kostner 3, Kristyan B Guppy-Coles 4, John J Atherton 4 and Chamindie Punyadeera 1,* 1 Saliva and Liquid Biopsy Translational Research Team, School of Biomedical Sciences, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, Queensland 4059, Australia; [email protected] 2 School of Biomedical Sciences, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, Queensland 4059, Australia; [email protected] 3 Department of Cardiology, Mater Adult Hospital, Brisbane, Queensland 4101, Australia; [email protected] 4 Cardiology Department, Royal Brisbane and Women’s Hospital and University of Queensland School of Medicine, Brisbane, Queensland 4029, Australia; [email protected] (K.B.G.-C.); [email protected] (J.J.A.) * Correspondence: [email protected]; Tel.: +61-7-3138-0830 Received: 22 October 2019; Accepted: 19 November 2019; Published: 22 November 2019 Abstract: Screening for systolic heart failure (SHF) has been problematic. Heart failure management guidelines suggest screening for structural heart disease and SHF prevention strategies should be a top priority. We developed a multi-protein biomarker panel using saliva as a diagnostic medium to discriminate SHF patients and healthy controls. We collected saliva samples from healthy controls (n = 88) and from SHF patients (n = 100). We developed enzyme linked immunosorbent assays to quantify three specific proteins/peptide (Kallikrein-1, Protein S100-A7, and Cathelicidin antimicrobial peptide) in saliva samples. The analytical and clinical performances and predictive value of the proteins were evaluated.