Sequence and Evolution of HLA-DR7- and -Drw53-Associated ,8-Chain Genes (Class II Histocompatibility Antigens/Gene Conversion) JOHN A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bilag 1 Output for Terror/Radikaliseringsbevillingen 2013 Uddannelses- Og Forskningsudvalget 2013-14 FIV Alm.Del Bilag 249 Offe
Uddannelses- og Forskningsudvalget 2013-14 FIV Alm.del Bilag 249 Offentligt Bilag 1 Output for terror/radikaliseringsbevillingen 2013 POLICY BRIEFING & SEMINARER, OPLÆG, VIDENSKABELIGE AVIS- INTERVIEW CENSOR RAPPORT RÅGIVNING KONFERENCER FOREDRAG & ARTIKLER, ARTIKLER REFEREE ER & WORKSHOPS FORELÆSNINGER BOGKAPITLER & COMMENTS 4 23 17 63 16 11 436 7 POLICY RAPPORTER Titel Forfatter DIIS rapport nr. Bottom-up Pakistan: Bringing context Mona Kanwal Sheikh 2013:1 and local culture into aid thinking Engaging with islamists: A new agenda Mona Kanwal Sheikh 2013:1 for the policy community Taliban talks: Past present and Mona Kanwal Sheikh 2013:6 prospects for the US, Afghanistan and & Maja Touzari Janesdatter Pakistan Greenwood (Red) Militante islamistiske grupper i Mali: Manni Crone 2013:8 ideologi, strategi og alliancer BRIEFING & RÅGIVNING Sted/Modtager Oplægsholder Dato/Opgave Hærens Operative Kommando, Manni Crone 07.02.2013 Gidseltagninger i Sahel UM Mona Kanwal Sheikh 16.04.2013 Briefing ny ambassadør til Pakistan PET og Social- og Ann-Sophie Hemmingsen 29.04.2013 Integrationsministeriet Oplæg om militant islamisme på Grunduddannelsesforløb for nøglepersoner ved kommuner og politi. Forebyggelse af radikalisering og ekstremisme 1 Social- og Integrationsministeriet Ann-Sophie Hemmingsen 12.04.2013 Briefing om online radikalisering UM’s MENA-kontor Rasmus Alenius Boserup 09.07.2013 Egypten UM’s MENA-team inklusiv ny Rasmus Alenius Boserup 09.07.2013 MENA-chef Egypten Ambassadør Pernille Kardel (Cairo) Rasmus Alenius Boserup 06.09.2013 Situationen -

Kanaloversigt - Digital Tv Gældende for Eniig, Nord Energi Og RAH Fibernet
Kanaloversigt - Digital tv Gældende for Eniig, Nord Energi og RAH fibernet Plads Navn Tv-Pakke Plads Navn Tv-Pakke Lille Mellem Stor Lille Mellem Stor 1 DR 1 X 39 Folketinget X TV 2 | Danmark ID Investigation 2 X 40 X HD Discovery TV2 Regional TV3 3 X 41 Viasat Explore HD X HD Danmark 4 TV3+ HD X 42 Viasat History HD X 5 Kanal 5 X 43 Viasat Nature HD X 6 6’eren HD X 44 V Sport Golf X 7 Kanal 4 X 45 CNN X 8 TV3 Puls HD X 46 TV 2 Sport X X 9 TV 2 Charlie X 47 Boomerang X 10 TV 2 Zulu X 48 Cartoon Network X 11 TV 2 News X 50 Disney Junior X 12 TV 2 Sport X 51 Nick Jr X 13 TV 2 Fri X 54 V Sport Ultra HD X 14 TV3 SPORT X 55 NatGeo Wild HD X 15 TV3 MAX X 62 TV2 | Østjylland X 16 DK4 X 63 TV/Midtvest X Tv Midtvest 17 Ekstrakanalen X 64 TV2/Nord Salto X Tv2 Nord Salto Discovery 18 X 65 TV SYD+ X Channel HD Tv Syd National 70 Østjylland X 19 X Geographic Kanal Østjylland 71 Midt/vest X 20 Animal Planet HD X Kanal Midtvest 72 Kanal Nord X 21 TLC HD X Kanal Nord 73 Syd X 22 Nickelodeon HD X Kanal Syd 78 VH1 X 23 Disney Channel X VH1 85 SVT1 HD X Paramount 24 X SVT 1 Network 87 TV4 Sverige HD X 25 CANAL9 HD X TV4 Sverige 88 DR1Syn X 26 Eurosport HD X DR 1Synstolkning 89 DR2Syn X 27 Eurosport 2 HD X DR 2 Synstolkning 91 NRK 1 HD X 28 MTV HD X NRK 1 93 TV 2 Norge HD X 29 Xee X TV2 Norge 100 Das Erste HD X TV 2 | Danmark Das Erste 30 X HD (Nord) 101 ZDF HD X TV 2 | Danmark ZDF 31 X HD (Østjylland) 103 Sat1 X Sat 1 TV 2 | Danmark 32 X HD (Syd) 104 RTL HD X RTL 33 DR 2 X 153 Stofa Kanalen X Stofa Kanalen 36 DR Ramasjang X Kanaloversigt - Digital radio -

Kammeradvokaten: Tidligere Banedanmark-Direktør Misbrugte Ikke Embedet | Nyheder | DR 31/07/2016 01.51
Kammeradvokaten: Tidligere Banedanmark-direktør misbrugte ikke embedet | Nyheder | DR 31/07/2016 01.51 NYHEDER TV RADIO MERE Privatlivspolitik Søg på dr.dk ) Nyhedsoverblik Politik Sport Viden Kultur Debat Lev Nu Regionalt Vejr Alle nyheder NYHEDER Juncker: A!ale mellem EU Iranske mænd iklæder sig CIA-chef tror ikke Syrien og Tyrkiet bliver stadig hijab i protest og solidaritet forbliver et samlet land ← mere skrøbelig 30. JUL. 2016 KL. 19.00 30. JUL. 2016 KL. 21.00 → 30. JUL. 2016 KL. 22.30 29. MAR. 2016 KL. 14.28 TOPHISTORIER Kammeradvokaten: Tidligere Banedanmark- direktør misbrugte ikke embedet Det var ikke en ordre, da Banedanmark-direktøren på et møde udtalte sig kritisk om et andet entreprenørfirma. Juncker: A!ale mellem EU og Tyrkiet bliver stadig mere skrøbelig Iranske mænd iklæder sigCIA-chef tror ikke Syrien hijab i protest og forbliver et samlet land solidaritet SENESTE INDLAND ' Banker kan presses til billigere boliglån 30. JUL. 2016 KL. 23.09 Banedanmarks tidligere direktør Jesper Hansen kom med en følelsesmæssig bemærkning om Ravn Bane Aps, men det var ikke en ordre om sortlistning, siger Kammeradvokaten. (Foto: Emil Hougaard © Scanpix) ' Studieoptag: Tusindvis af unge ringer og skriver til national rådgivning 30. JUL. 2016 KL. 17.50 ⎙ PRINT Af Emma To! ' Stor forvirring: Standby-plads blev vekslet til DEL ARTIKLEN: studieplads - og om igen Banedanmarks tidligere administrerende direktør Jesper Hansen $ MAIL 30. JUL. 2016 KL. 16.30 bliver nu renset for beskyldninger om embedsmisbrug. % TWITTER Det fremgår af en undersøgelse, som Kammeradvokaten har lavet ALLE INDLAND & FACEBOOK for Transport- og Bygningsministeriet, efter at direktøren i 'Ravn Bane Aps', Jesper Ravn, politianmeldte Banedanmark-direktøren i starten af året. -

Pressemeddelelse (Gallup Radio-Meter) 13
Pressemeddelelse (Gallup Radio-Meter) 13. marts 2018 Lyttertal uge 5 (29. januar - 4. februar) Eftertryk/citat må kun ske mod flg. kildeangivelse: "Kantar Gallup Radio-Meter" Kantar Gallup måler hvert døgn på vegne af den danske radiobranche radio lytningen i Danmark ved hjælp af Radio metre (PPM'er)*. Tallene i denne pressemeddelelse er opgivet for hele befolkningen (12 år og derover) svarende til 4.987.000 personer, og endvidere i målgruppen 15-50 år, svarende til 2.641.000 personer. Hvor meget radio lytter danskerne til? I uge 5 lyttede en gennemsnitsdansker på 12 år og derover i gennemsnit til de stationer, der er med i denne undersøgelse i 12 timer og 53 minutter, hvoraf 10 timer og 41 minutter fandt sted i perioden 06.00-18.00. Ugentlig Ugentlig Ugentlig Ugentlig Målgruppe dækning dækning Målgruppe dækning dækning 12-50 år i 1.000 i % Share i % 12 år+ i 1.000 i % Share i % DR P3 1.283 48,7 23,7 DR P41 2.891 58,0 39,1 DR P41 1.258 47,8 26,0 DR P3 1.907 38,2 11,5 NOVA 868 33,0 7,2 NOVA 1.162 23,3 3,7 The Voice2 477 18,1 3,9 DR P1 794 15,9 7,6 Radio 100 326 12,4 1,6 The Voice2 654 13,1 1,9 Pop FM 304 11,6 3,6 DR P5 546 10,9 7,9 DR P1 282 10,7 2,7 DR P7 Mix 482 9,7 1,3 DR P7 Mix 273 10,4 2,0 Radio24syv 481 9,6 3,1 Skala FM 270 10,3 4,5 Pop FM 478 9,6 1,8 Radio Soft 217 8,3 2,4 Radio 100 417 8,4 0,8 Radio24syv 192 7,3 1,4 Radio Soft 372 7,5 1,3 VLR8 173 6,6 1,4 Skala FM 359 7,2 2,2 myRock10 122 4,6 1,2 DR P2 352 7,1 2,9 Classic FM Total14 118 4,5 0,6 VLR8 307 6,2 1,5 DR P5 97 3,7 1,0 Classic FM Total14 175 3,5 0,4 DR P2 69 2,6 0,2 myRock10 -

Pressemeddelelse (Gallup Radio-Meter) 7
Pressemeddelelse (Gallup Radio-Meter) 7. december 2011 Lyttertal uge 48 (28. november - 4. december) Eftertryk/citat må kun ske mod flg. kildeangivelse: "TNS Gallup Radio-Meter" TNS Gallup måler hvert døgn på vegne af den danske radiobranche radio lytningen i Danmark ved hjælp af Radio metre (PPM'er)*. Tallene i denne pressemeddelelse er opgivet for hele befolkningen (12 år og derover) svarende til 4.717.000 personer, og endvidere i målgruppen 12-50 år, svarende til 2.837.000 personer. Hvor meget radio lytter danskerne til? I uge 48 lyttede en gennemsnitsdansker på 12 år og derover i gennemsnit til de stationer, der er med i denne undersøgelse i 14 timer og 26 minutter, hvoraf 12 timer og 8 minutter fandt sted i perioden 06.00-18.00. Ugentlig Ugentlig Ugentlig Ugentlig Målgruppe dækning dækning Målgruppe dækning dækning 12-50 år i 1.000 i % Share i % 12 år+ i 1.000 i % Share i % DR P3 1.701 60,0 29,5 DR P4 Total1 2.798 59,3 44,0 DR P4 Total1 1.401 49,5 28,3 DR P3 2.384 50,5 17,9 NOVA fm 1.118 39,5 7,5 NOVA fm 1.430 30,3 4,5 The Voice Total2 680 24,0 4,1 The Voice Total2 879 18,6 2,1 Radio 100 459 16,2 4,2 DR P1 761 16,1 7,1 Pop FM 446 15,7 2,5 Pop FM 736 15,6 2,8 Radio Soft 344 12,1 3,4 Radio 100 558 11,8 2,2 DR P7 Mix 338 11,9 2,7 DR P7 Mix 507 10,7 1,5 DR P1 269 9,5 1,6 Radio Soft 494 10,5 1,8 DR Mama 164 5,8 0,6 DR P2 Klassisk 427 9,0 2,4 Radio24syv 145 5,1 1,2 Radio24syv 380 8,1 1,9 DR P5 60 2,1 0,4 DR Mama 221 4,7 0,3 DR P2 Klassisk 58 2,1 0,2 DR P5 212 4,5 1,6 go!FM 51 1,8 0,1 DR Nyheder 77 1,6 0,2 DR Ramasjang Radio 50 1,8 0,1 DR P6 Beat 73 1,5 0,1 Grupper Grupper Public Service7 2.366 83,5 64,9 Public Service7 4.105 87,0 77,2 DR Total3 2.355 83,1 63,7 DR Total3 4.077 86,4 75,3 Kommercielle stationer4 2.284 80,6 35,1 Kommercielle stationer4 3.399 72,1 22,8 SBS Radio Total5 1.634 57,7 16,0 DDR Total6 2.375 50,4 12,2 DDR Total6 1.631 57,6 19,0 SBS Radio Total5 2.284 48,4 10,4 Stationerne er sorteret efter ugentlig dækning i 1.000. -
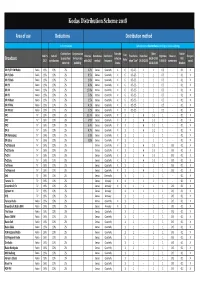
Distribution Scheme 2018
Kodas Distribution Scheme 2018 Area of use Deductions Distribution method % of net revenue Calculated on a duration basis according to below weightings Contributions Compensation Extended Other Station Adm. % Cultural Revenue Distribution Distribution Live Time factor- Primetime Nighttime Music in First perf. to subsidised for local sub- collective 06.00-19.30 ID's/ Broadcast 2017 contributions splits 2017 method frequence factor when "Live" 19.30-22.30 0.00-6.00 commercials award concert ¤) publshing license 22.30-23.59 Breakers DR P1/P2 FM Radio Radio 16% 10% 2% 1.2% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P1 Dab Radio 16% 10% 2% 0.5% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P2 Dab Radio 16% 10% 2% 1.0% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P3 Radio 16% 10% 2% 8.0% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P4 Radio 16% 10% 2% 15.0% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P5 Radio 16% 10% 2% 5.0% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P6 Beat Radio 16% 10% 2% 3.5% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P7 Mix Radio 16% 10% 2% 4.1% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR P8 Jazz Radio 16% 10% 2% 3.2% Census Quarterly X 6 0.5-2.5 1 1 0.5 - 0.1 X DR1 TV 16% 10% 2% 32.7% Census Quarterly X 1 1 6 1-2 1 - 0.1 X DR2 TV 16% 10% 2% 5.8% Census Quarterly X 1 1 6 1-2 1 - 0.1 X DR3 TV 16% 10% 2% 6.4% Census Quarterly X 1 1 6 1-2 1 - 0.1 X DR K TV 16% 10% 2% 4.7% Census Quarterly X 1 1 6 1-2 1 - 0.1 X DR Ramasjang TV 16% 10% 2% 5.0% Census Quarterly X 1 1 1 1 1 - 0.1 X DR Ultra TV 16% -

Evolutionary and Genetic Implications of Sequence Variation In
Proc. Natl. Acad. Sci. USA Vol. 84, pp. 209-213, January 1987 Immunology Evolutionary and genetic implications of sequence variation in two nonallelic HLA-DR 13-chain cDNA sequences (HLA class II antigens/genetic polymorphism/DNA sequence homology) JULIE MISKIMEN CURTSINGER, JOANNE M. HILDEN, J. SCOTT CAIRNS, AND FRITZ H. BACH Immunobiology Research Center, Department of Laboratory Medicine and Pathology, Box 724 Mayo Building, University of Minnesota, Minneapolis, MN 55455 Communicated by Donald C. Shreffler, August 25, 1986 ABSTRACT Most HLA haplotypes carry two expressed cells that are not homozygous for DR have made it difficult DR 13-chain genes; in the DR4 haplotype, the polymorphic locus to assign a DNA sequence to a particular locus within the DR has been called DR p13 and the apparently nonpolymorphic family and to determine to what degree the variation seen locus has been called DR 12. We have isolated nearly full-length among DR 13 sequences represents allelic variation versus DR 8-chain cDNA clones representing each of these two loci interlocus (isotypic) differences. To address these questions from a cell line homozygous for DR4 and Dw4. The clones have we report here a sequence ofa nearly full-length DR P2 cDNA been sequenced and the sequences compared with published clone derived from a DR4 homozygous cell, which allows DR 13 cDNA sequences derived from other haplotypes. A sequence comparisons to be made with known DR ,1 comparison of our sequences with other published cDNA sequences of DR4 and with DR 13 sequences from other sequences did not allow assignment of these other sequences to haplotypes. -

DR Symfoniorkestret DR Koncertkoret Sarah Hicks, Dirigent Tuva
Torsdag 4. januar 2018 kl. 20.00 Fredag 5. januar 2018 kl. 18.30 & 21.00 DR SymfoniOrkestret DR Koncertkoret Sarah Hicks, dirigent Tuva Semmingsen, mezzosopran Hans Ulrik, saxofon Kristian Leth, vært Duellen – Morricone trækker først DR Koncerthuset 2017/18 2 Program Torsdag 4. januar kl. 20.00 Koncerten kan opleves på Fredag 5. januar kl. 18.30 Fredag 5. januar kl. 21.00 DR Koncerthuset fredag 5. januar koncertsalen kl. 19.20 DR SymfoniOrkestret DR KoncertKoret fredag 26. januar Sarah Hicks kl. 20.00 Dirigent Koncerten vil desuden blive tilgængelig på Edward Ananian-Cooper dr.dk/tv fra fredag 26. januar. korsyngemester Del dine oplevelser. Christine Nonbo Andersen #Duellen sopran #DRKoncerthuset Tuva Semmingsen mezzosopran Hans Ulrik saxofon, mundharpe, blokfløjter Mads Kjølby guitarer, banjo, mandolin Kristian Leth vært Christina Åstrand koncertmester Chef for DR Kor & Orkestre: Kim Bohr Producer: Thore Brinkmann Repertoirechef: Tatjana Kandel Musikteknik: Mikkel Nymand Kunstnerisk leder for DR KoncertKoret: Michael Emery Produktionsledere: Signe Møldrup, Producent: Nicolai Abrahamsen Jakob Helmer Mørck, Cecilie Honoré Duellen – Morricone trækker først DR Koncerthuset 2017/18 3 Program The Big Gundown (1966) & Inglourious Basterds (2009) DOPO LA CONDANNA Ennio Morricone (født 1928), arr. Martin Nygård Jørgensen A Fistful of Dollars (1964) A FISTFUL OF DOLLARS Ennio Morricone, arr. Thommy Andersson The Hateful Eight (2015) OUVERTURE Ennio Morricone, arr. Martin Nygård Jørgensen Once Upon a Time in the West (1968) SUITE: MAN WITH A HARMONICA, CHEYENNE, MAIN THEME Ennio Morricone, arr. Matthias Keller/Mike Cholewa Solister: Hans Ulrik & Tuva Semmingsen Taxi Driver (1976) TAXI DRIVER - A NIGHT PIECE Berrnard Herrmann (1911-1975), arr. Christopher Palmer Solist: Hans Ulrik The Untouchables (1987) THE STRENGTH OF THE RIGHTEOUS Ennio Morricone, arr. -

11. September 2018 Årets Nomineringer Til Prix Radio
PRESSEMEDDELSE – 11. september 2018 Årets nomineringer til Prix Radio Fagjuryerne har haft travlt. 297 indstillede bidrag er blevet lyttet igennem og reduceret til 54 nomineringer. Antallet af indstillede er steget år efter år og Prix Radio kunne i år melde om rekord med næsten 300 indstillede bidrag. Juryerne har været imponerede over den kvalitet og kreativitet, der kommer til udtryk i de mange indstillede bidrag. Men allermest har juryerne gang på gang udtrykt respekt og anerkendelse for det håndværk, som den danske radiobranche kan præstere. Hvad enten der er tale om den lille freelance produktion, der med de midler, der er til rådighed energisk, dygtigt og med en imponerende fortællelyst præsenterer lytterne for alt fra musikprogrammer til dokumentar, feature og interviewhistorier til de store, åbenlyst ressourcekrævende produktioner, der virkelig udnytter alle de muligheder de bliver givet til glæde og oplysning for lytterne. Jurymedlem gennem mange år Tina Bilsbo siger om juryarbejdet og de nominerede: ”Juryarbejdet kombinerer og aktiverer nysgerrigheden, fagligheden og passionen når de mange nominerede lyttes til og diskuteres. Der er en utrolig høj standard og alsidighed i den radio vi som jury er blevet præsenteret for i år, det både udfordrer og glæder mine øre - og mit radiohjerte!” Brugerredaktør Tom Bue fra Nordjyske Medier, der ligeledes har siddet i juryen i mange år fortsætter: ”Jeg bliver glad helt ind i mit radiohjerte, når jeg hører hvor meget god radio der bliver lavet i Danmark – indenfor alle genrer er standarden virkelig høj!” Og endelig bemærker Anne Sophia Hermansen, der er ny i juryen, at: ”Radio er det nye sort, og den tid er forbi, hvor radio var synonymt med den gamle verden. -

Drs Public Service-Redegørelse 2012
DRs public service-redegørelse 2012 UDGIVET 06.05.2013 AF DR DESIGN: DR DESIGN TrYK: ROSENDAHL SCHULTZ A/S s04 0 Indledning s05 1 Fordeling af programtyper på tv- og radiokanaler s11 2 Genudsendelse af programmer s12 3 Danskernes forbrug af DRs programudbud s14 4 Danskernes vurdering af DRs indholdskvalitet s21 5 DRs indhold på internettet mv s23 6 DRs regionale radioprogramvirksomhed s25 7 Nyheder s27 8 Uddannelse og læring s29 9 Børn og beskyttelse af børn s32 10 Unge s34 11 Dansk drama s36 12 Dansk musik s39 13 Dansk kultur s42 14 Smalle idrætsgrene og handicapidræt s44 15 Hensyn til handicappede s49 16 Dansksprogede programmer og sprogpolitik s53 17 Europæiske tv-programmer og europæiske programmer fra uafhængige producenter s55 18 Støtte til dansk film s56 19 Udlægning af produktion s59 20 Dialog med danskerne Indhold 0 Indledning Public service-kontraktens krav til princip er blandt andet indført, fordi der DRs redegørelse på tv er sket en udvikling i måden at bruge Public service-kontrakten mellem DR og genudsendelser på, og for at sikre, at DR har kulturministeren for perioden 1. januar 2011 fleksibilitet i planlægningen af indholdet på til 31. december 2014 fastlægger, hvilke tv-kanalerne. I afsnittet om genudsendelser opgaver DR skal opfylde og udstikker ram- findes til gengæld en grundig generel beskri- merne for DRs public service-virksomhed. velse af DRs brug af genudsendelser. Det fremgår af kontrakten, at DR årligt skal DR angiver som et princip niveauer for det udarbejde en redegørelse for, hvordan public år, som redegørelsen omhandler samt for de service-forpligtelserne er blevet opfyldt i det tidligere år i den aktuelle kontraktperiode. -

Change of PID Values for Services on Thor 5, Transponder BSS37
Fornebu, 28th November 2013 Change of PID values for services on Thor 5, Transponder BSS37. As the services on Thor 5, transponder BSS37, will be moved to a new hardware platform, 4 th December at 10:00 CET, some of the services will be assigned new PID values. Please note that the Service ID will not be changed. Transponder parameters (no changes): Satellite: Thor 5 Transponder: BSS37 Downlink frequency: 12418 MHz Downlink polarisation: Vertical Symbol rate: 28,0 Msym/sec Forward Error Correction: 7/8 DVB-S, QPSK The changes will be: AlJazeera English: CCTV News : Video PID 1186 Video PID 1166 Audio PID 3182 Audio PID 3097 PMT PID 214 PMT PID 495 CCTV 9 Documentary: Scandinavian Satellite Radio: Video PID 1172 Audio PID 3174 Audio PID 3102 PMT PID 128 PCR PID 8188 PMT PID 532 BrunstadTV: France24: Video PID 1102 Video PID 1101 Audio PID 3101 (nor) Audio PID 3072 Audio PID 3315 (deu) PMT PID 104 Audio PID 3364 (dut) ECM PID 7134 (Conax Canal Digital 0x0B00) Audio PID 3373 (eng) ECM PID 7135 (UPC Nagra 0x1815) Audio PID 4301 (fre) ECM PID 7136 (UPC Cryptoworks 0x0D02) Audio PID 4302 (spa) ECM PID 7137 (UPC Cryptoworks 0x0D97) Audio PID 4303 (fin) ECM PID 7138 (UPC Irdeto 0x0653) Audio PID 4304 (rus) ECM PID 7139 (UPC Conax 0x0B02) Audio PID 4305 (por) Audio PID 4306 (rum) Audio PID 4307 (tur) Audio PID 4308 (bul) Audio PID 4309 (pol) Audio PID 4311 (hun) Audio PID 4312 (ita) Audio PID 4313 (cze) DVBSubtitle PID 6672 (nor) DVBSubtitle PID 6673 (deu) DVBSubtitle PID 6674 (dut) DVBSubtitle PID 6675 (eng) ECM PID 7090 PMT PID 346 EMM PID 49 Services in Transponder BSS37 with no PID changes: BBC Knowledge Sky News DR1 Syd & DR2 Syd DR P1, DR P2, DR P3, DR P4 København & DR P5 Svein Bråten Senior CSM – DTH Nordic Broadcasting Systems - Technology Division Telenor Satellite Broadcasting . -

Priser Til Dr 2018
PRISER TIL DR 2018 PRISER TIL DR TV Januar Nordisk Film- og TV-Fonds Pris for fremragende tv-drama til hovedforfatter Adam Price for dramaserien ’Herrens Veje’ To priser til DR ved Zulu Awards 2018: • Årets tv-vært: Sofie Linde • Årets originale tv-program: ’Petra elsker sig selv’, DR3 Tre priser til DR-programmer ved Reality Awards 2018: • Årets underholdningsprogram: ’Den Store Bagedyst’, DR1 • Årets bemærkning og årets par: ’En sugardaters bekendelser’, DR3 11 priser til DR-programmer og DR-programmedarbejdere ved uddelingen af TV-Prisen 2018: • Bedste børneprogram: ’Klassen’, DR Danmark B&U for DR Ultra • Bedste crime: ’Morderen på den hvide hest’, DR Ung for DR3 • Bedste dokumentar: ’De sidste mænd i Aleppo’, Larm film/Aleppo Media Center, Kloos & Co. Medien for DR2 • Bedste faktaserie: ’De skjulte talenter’, DR Danmark og DR Dokumentar for DR1 • Bedste karakterdrevne serie: ’Prinsesser fra Blokken’, DR Ung for DR3 • Bedste reportageserie: ’Gina Jaqueline’, Pineapple Entertainment for DR3 • Bedste satire/comedy: ’Tæt på sandheden’, Small Shoes for DR2 • Bedste TV-serie: ’Herrens Veje’, DR Drama for DR1 • Årets nyskabelse – fakta: ’Historien om Danmark’, DR Kultur for DR1 • Seerprisen: ’Historien om Danmark’, DR Kultur for DR1 • Årets Otto: Dokumentaristen Poul-Erik Heilbuth, DR Februar Fem Robert-priser til DRs dramaserier ved Danmarks Film Akademis uddeling af film- og tv-Robert’er 2018 • Årets tv-serie: ’Herrens Veje’ • Årets mandlige hovedrolle – tv-serie: Lars Mikkelsen i ’Herrens Veje’ • Årets kvindelige hovedrolle – tv-serie: Ann Eleonora Jørgensen i ’Herrens Veje’ • Årets mandlige birolle – tv-serie: Mikkel Boe Følsgaard i ’Arvingerne III’ • Årets kvindelige birolle – tv-serie: Lene Maria Christensen i ’Arvingerne III’ • Hædersbuket fra KLF, Kirke & Medier til redaktionen bag forbrugermagasinet ’Kontant’ på DR1 for at sætte fokus på god etik.