Symbolic Proof of Bistability in Reaction Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Geometry of Convex Cones Associated with the Lyapunov Inequality and the Common Lyapunov Function Problem∗
Electronic Journal of Linear Algebra ISSN 1081-3810 A publication of the International Linear Algebra Society Volume 12, pp. 42-63, March 2005 ELA www.math.technion.ac.il/iic/ela THE GEOMETRY OF CONVEX CONES ASSOCIATED WITH THE LYAPUNOV INEQUALITY AND THE COMMON LYAPUNOV FUNCTION PROBLEM∗ OLIVER MASON† AND ROBERT SHORTEN‡ Abstract. In this paper, the structure of several convex cones that arise in the studyof Lyapunov functions is investigated. In particular, the cones associated with quadratic Lyapunov functions for both linear and non-linear systems are considered, as well as cones that arise in connection with diagonal and linear copositive Lyapunov functions for positive linear systems. In each of these cases, some technical results are presented on the structure of individual cones and it is shown how these insights can lead to new results on the problem of common Lyapunov function existence. Key words. Lyapunov functions and stability, Convex cones, Matrix equations. AMS subject classifications. 37B25, 47L07, 39B42. 1. Introduction and motivation. Recently, there has been considerable inter- est across the mathematics, computer science, and control engineering communities in the analysis and design of so-called hybrid dynamical systems [7,15,19,22,23]. Roughly speaking, a hybrid system is one whose behaviour can be described math- ematically by combining classical differential/difference equations with some logic based switching mechanism or rule. These systems arise in a wide variety of engineer- ing applications with examples occurring in the aircraft, automotive and communi- cations industries. In spite of the attention that hybrid systems have received in the recent past, important aspects of their behaviour are not yet completely understood. -
![Arxiv:1909.13402V1 [Math.CA] 30 Sep 2019 Routh-Hurwitz Array [14], Argument Principle [23] and So On](https://docslib.b-cdn.net/cover/6437/arxiv-1909-13402v1-math-ca-30-sep-2019-routh-hurwitz-array-14-argument-principle-23-and-so-on-676437.webp)
Arxiv:1909.13402V1 [Math.CA] 30 Sep 2019 Routh-Hurwitz Array [14], Argument Principle [23] and So On
ON GENERALIZATION OF CLASSICAL HURWITZ STABILITY CRITERIA FOR MATRIX POLYNOMIALS XUZHOU ZHAN AND ALEXANDER DYACHENKO Abstract. In this paper, we associate a class of Hurwitz matrix polynomi- als with Stieltjes positive definite matrix sequences. This connection leads to an extension of two classical criteria of Hurwitz stability for real polynomials to matrix polynomials: tests for Hurwitz stability via positive definiteness of block-Hankel matrices built from matricial Markov parameters and via matricial Stieltjes continued fractions. We obtain further conditions for Hurwitz stability in terms of block-Hankel minors and quasiminors, which may be viewed as a weak version of the total positivity criterion. Keywords: Hurwitz stability, matrix polynomials, total positivity, Markov parameters, Hankel matrices, Stieltjes positive definite sequences, quasiminors 1. Introduction Consider a high-order differential system (n) (n−1) A0y (t) + A1y (t) + ··· + Any(t) = u(t); where A0;:::;An are complex matrices, y(t) is the output vector and u(t) denotes the control input vector. The asymptotic stability of such a system is determined by the Hurwitz stability of its characteristic matrix polynomial n n−1 F (z) = A0z + A1z + ··· + An; or to say, by that all roots of det F (z) lie in the open left half-plane <z < 0. Many algebraic techniques are developed for testing the Hurwitz stability of matrix polynomials, which allow to avoid computing the determinant and zeros: LMI approach [20, 21, 27, 28], the Anderson-Jury Bezoutian [29, 30], matrix Cauchy indices [6], lossless positive real property [4], block Hurwitz matrix [25], extended arXiv:1909.13402v1 [math.CA] 30 Sep 2019 Routh-Hurwitz array [14], argument principle [23] and so on. -

Non-Hurwitz Classical Groups
London Mathematical Society ISSN 1461–1570 NON-HURWITZ CLASSICAL GROUPS R. VINCENT and A.E. ZALESSKI Abstract In previous work by Di Martino, Tamburini and Zalesski [Comm. Algebra 28 (2000) 5383–5404] it is shown that cer- tain low-dimensional classical groups over finite fields are not Hurwitz. In this paper the list is extended by adding the spe- cial linear and special unitary groups in dimensions 8,9,11,13. We also show that all groups Sp(n, q) are not Hurwitz for q even and n =6, 8, 12, 16. In the range 11 <n<32 many of these groups are shown to be non-Hurwitz. In addition, we observe that PSp(6, 3), P Ω±(8, 3k), P Ω±(10,q), Ω(11, 3k), Ω±(14, 3k), Ω±(16, 7k), Ω(n, 7k) for n =9, 11, 13, PSp(8, 7k) are not Hurwitz. 1. Introduction A finite group H = 1 is called Hurwitz if it is generated by two elements X, Y satisfying the conditions X2 = Y 3 =(XY )7 = 1. A long-standing problem is that of classifying simple Hurwitz groups. The problem has been solved for alternating groups by Conder [4], and for sporadic groups by several authors with the latest result by Wilson [27]. It remains open for groups of Lie type and for classical groups. 3 2 Quite a lot is known. Groups D4(q) for (q, 3)=1, G2(q),G2(q) are Hurwitz with 3 k few exceptions, groups D4(3 ) are not Hurwitz; see Jones [10] and Malle [15, 16]. -

Analysis and Control of Linear Time–Varying Systems Exercise 2.3
Analysis and control of linear time–varying 2 systems Before considering the actual subject matter, i.e., analysis and control design of nonlinear systems, at first linear time-varying systems or LTV systems of the form x˙ A(t)x B(t)u, t t , x(t ) x (2.1a) Æ Å È 0 0 Æ 0 y C(t)x D(t)u, t t (2.1b) Æ Å ¸ 0 with x(t) Rn, u(t) Rm, y(t) Rp and 2 2 2 n n n m p n p m A(t): RÅ R £ , B(t): RÅ R £ , C(t): RÅ R £ , D(t): RÅ R £ t 0 ! t 0 ! t 0 ! t 0 ! are considered. Here RÅ : {t R t t 0} denotes the set of real numbers larger or equal to t0. t 0 Æ 2 j ¸ The motivation behind this sequential approach is that LTV systems on the one hand show certain similarities to linear time–invariant systems but are distinguished by significant differences in their dynamic analysis. In this sense, they — at least to some extend — resemble nonlinear systems. On the other hand, the linearization of a nonlinear system around a solution trajectory t (x¤(t),u¤(t)) 7! yields an LTV system. Hence the subsequently introduced tools can be used for control and observer design at least in a neighborhood of the trajectory (x¤(t),u¤(t)). 2.1 Transition matrix and solution of the state differential equations At first free or autonomous systems x˙ A(t)x, t t , x(t ) x (2.2) Æ È 0 0 Æ 0 are addressed. -

Learning Stable Dynamical Systems Using Contraction Theory
Learning Stable Dynamical Systems using Contraction Theory eingereichte MASTERARBEIT von cand. ing. Caroline Blocher geb. am 19.08.1990 wohnhaft in: Leonrodstrasse 72 80636 M¨unchen Tel.: 0176 27250499 Lehrstuhl f¨ur STEUERUNGS- und REGELUNGSTECHNIK Technische Universit¨atM¨unchen Univ.-Prof. Dr.-Ing./Univ. Tokio Martin Buss Fachgebiet f¨ur DYNAMISCHE MENSCH-ROBOTER-INTERAKTION f¨ur AUTOMATISIERUNGSTECHNIK Technische Universit¨atM¨unchen Prof. Dongheui Lee, Ph.D. Betreuer: M.Sc. Matteo Saveriano Beginn: 03.08.2015 Zwischenbericht: 25.09.2015 Abgabe: 19.01.2016 In your final hardback copy, replace this page with the signed exercise sheet. Abstract This report discusses the learning of robot motion via non-linear dynamical systems and Gaussian Mixture Models while optimizing the trade-off between global stability and accurate reproduction. Contrary to related work, the approach used in this thesis seeks to guarantee the stability via Contraction Theory. This point of view allows the use of results in robust control theory and switched linear systems for the analysis of the global stability of the dynamical system. Furthermore, a modification of existing approaches to learn a globally stable system and an approach to locally stabilize an already learned system are proposed. Both approaches are based on Contraction Theory and are compared to existing methods. Zusammenfassung Diese Arbeit behandelt das Lernen von stabilen dynamischen Systemen ¨uber eine Gauss’sche Mischverteilung. Im Gegensatz zu bisherigen Arbeiten wird die Sta- bilit¨at des Systems mit Hilfe der Contraction Theory untersucht. Ergebnisse aus der robusten Regulung und der Stabilit¨at von schaltenden Systemen k¨onnen so ¨ubernommen werden. Um die Stabilit¨at des dynamischen Systems zu garantieren und gleichzeitig die Bewegung des gelernten Systems m¨oglichst wenig zu beein- flussen, wird eine Anpassung der bereits bestehenden Methode an die Bewegung vorgeschlagen. -

Generalized Hurwitz Matrices, Generalized Euclidean Algorithm
Generalized Hurwitz matrices, generalized Euclidean algorithm, and forbidden sectors of the complex plane Olga Holtz∗†, Sergey Khrushchev,‡ and Olga Kushel§ June 23, 2015 Abstract Given a polynomial n n−1 f(x)= a0x + a1x + · · · + an with positive coefficients ak, and a positive integer M ≤ n, we define a(n infinite) generalized Hurwitz matrix HM (f):=(aMj−i)i,j . We prove that the polynomial f(z) does not vanish in the sector π z ∈ C : | arg(z)| < n M o whenever the matrix HM is totally nonnegative. This result generalizes the classical Hurwitz’ Theo- rem on stable polynomials (M = 2), the Aissen-Edrei-Schoenberg-Whitney theorem on polynomials with negative real roots (M = 1), and the Cowling-Thron theorem (M = n). In this connection, we also develop a generalization of the classical Euclidean algorithm, of independent interest per se. Introduction The problem of determining the number of zeros of a polynomial in a given region of the complex plane is very classical and goes back to Descartes, Gauss, Cauchy [4], Routh [22, 23], Hermite [9], Hurwitz [14], and many others. The entire second volume of the delightful Problems and Theorems in Analysis by P´olya and Szeg˝o[21] is devoted to this and related problems. See also comprehensive monographs of Marden [17], Obreshkoff [19], and Fisk [6]. One particularly famous late-19th-century result, which also has numerous applications, is the Routh- Hurwitz criterion of stability. Recall that a polynomial is called stable if all its zeros lie in the open left half-plane of the complex plane. The Routh-Hurwitz criterion asserts the following: Theorem 1 (Routh-Hurwitz [14, 22, 23]). -

Critical Exponents: Old and New
W&M ScholarWorks Undergraduate Honors Theses Theses, Dissertations, & Master Projects 5-2011 Critical Exponents: Old and New Olivia J. Walch College of William and Mary Follow this and additional works at: https://scholarworks.wm.edu/honorstheses Part of the Mathematics Commons Recommended Citation Walch, Olivia J., "Critical Exponents: Old and New" (2011). Undergraduate Honors Theses. Paper 423. https://scholarworks.wm.edu/honorstheses/423 This Honors Thesis is brought to you for free and open access by the Theses, Dissertations, & Master Projects at W&M ScholarWorks. It has been accepted for inclusion in Undergraduate Honors Theses by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. Critical exponents: Old and New A thesis submitted in partial fulfillment of the requirement for the degree of Bachelor of Science in Mathematics from The College of William and Mary by Olivia Walch Accepted for: Thesis director: CHARLES JOHNSON Panel Member 1: ILYA SPITKOVSKY Panel Member 2: JOSHUA ERLICH Williamsburg, VA May 13, 2011 Abstract Let P be a class of matrices, and let A be an m-by-n matrix in the class; consider some continuous powering, Aftg. The critical exponent of P, if it exists, with respect to the powering is the lowest power g(P) such that for any matrix B 2 P , Bftg 2 P 8 t > g(P). For powering relative to matrix multiplication in the traditional sense, hereafter referred to as conventional multiplication, t this means that A is in the specified class for all t > gC (P). For Hadamard (t) multiplication, similarly, A is in the class 8 t > gH (P). -

CONFERENCE PROGRAM Monday, April 23 09:30–10:30
CONFERENCE PROGRAM Monday, April 23 09:30–10:30 REGISTRATION 10:30–11:15 Pertti Mattila. Projections, Hausdorff dimension, and very little about analytic capacity. COFFEE BREAK 11:40–12:25 Arno Kuijlaars. The two-periodic Aztec diamond and matrix valued orthogonal polynomials. 12:35–13:20 Alexander Logunov. 0:01% improvement of the Liouville property for discrete harmonic functions on Z2. LUNCH Section A 15:00–15:25 Konstantin Dyakonov. Interpolating by functions from star-invariant subspaces. 15:30–15:55 Xavier Massaneda. Equidistribution and β-ensembles. Section B 15:00–15:25 Alexandre Sukhov. Analytic geometry of Levi-flat singularities. 15:30–15:55 Nikolai Kruzhilin. Proper maps of Reinhardt domains. COFFEE BREAK 1 Section A 16:25–16:50 St´ephaneCharpentier. Small Bergman–Orlicz and Hardy–Orlicz spaces. 16:55–17:20 Avner Kiro. Uniqueness theorems in non-isotropic Carleman classes. 17:25–17:50 Alexander Pushnitski. Weighted model spaces and Schmidt subspaces of Hankel operators. Section B 16:25–16:50 Vesna Todorˇcevi´c. Subharmonic behavior and quasiconformal mappings. 16:55–17:20 Nicola Arcozzi. Carleson measures for the Dirichlet spaces on the bidisc. 18:00 WELCOME PARTY at The Euler International Mathematical Institute 2 Tuesday, April 24 09:30–10:15 Armen Sergeev. Seiberg–Witten equations as a complex version of Ginzburg–Landau Equations. 10:25–11:10 Josip Globevnik. On complete complex hypersurfaces in CN . COFFEE BREAK 11:40–12:25 Alexander Aptekarev. Asymptotics of Hermite–Pad´e approximants of multi-valued analytic functions. 12:35–13:20 Frank Kutzschebauch. Factorization of holomorphic matrices in elementary factors. -

1 Definitions
Xu Chen February 19, 2021 Outline • Preliminaries • Stability concepts • Equilibrium point • Stability around an equilibrium point • Stability of LTI systems • Method of eigenvalue/pole locations • Routh Hurwitz criteria for CT and DT systems • Lyapunov’s direct approach 1 Definitions 1.1 Finite dimensional vector norms Let v 2 Rn. A norm is: • a metric in vector space: a function that assigns a real-valued length to each vector in a vector space, e.g., p T p 2 2 2 – 2 (Euclidean) norm: kvk2 = v v = v1 + v2 + ··· + vn Pn – 1 norm: kvk1 = i=1 jvjj (absolute column sum) – infinity norm: kvk1 = maxi jvij Pn p 1=p – p norm: jjvjjp = ( i=1 jvij ) ; 1 ≤ p < 1 • default in this set of notes: k · k = k · k2 1.2 Equilibrium state For n-th order unforced system x_ = f (x; t) ; x(t0) = x0 an equilibrium state/point xe is one such that f (xe; t) = 0; 8t • the condition must be satisfied by all t ≥ 0. • if a system starts at equilibrium state, it stays there • e.g., (inverted) pendulum resting at the verticle direction • without loss of generality, we assume the origin is an equilibrium point 1.3 Equilibrium state of a linear system For a linear system x_(t) = A(t)x(t); x(t0) = x0 • origin xe = 0 is always an equilibrium state • when A(t) is nonsingular, multiple equilibrium states exist 1 Xu Chen 1.4 Continuous function February 19, 2021 Figure 1: Continuous functions Figure 2: Uniformly continuous (left) and continuous but not uniformly continuous (right) functions 1.4 Continuous function The function f : R ! R is continuous at x0 if 8 > 0, there exists a δ (x0; ) > 0 such that jx − x0j < δ =) jf (x) − f (x0)j < Graphically, continuous functions is a single unbroken curve: 2 1 x2−1 e.g., sin x, x , x−1 , x−1 , sign(x − 1:5) 1.5 Uniformly continuous function The function f : R ! R is uniformly continuous if 8 > 0, there exists a δ () > 0 such that jx − x0j < δ () =) jf (x) − f (x0)j < • δ is a function of not x0 but only 1.6 Lyapunov’s definition of stability • Lyapunov invented his stability theory in 1892 in Russia. -
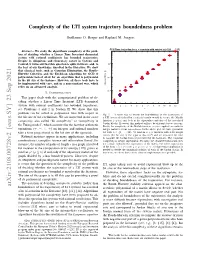
Complexity of the LTI System Trajectory Boundedness Problem
Complexity of the LTI system trajectory boundedness problem Guillaume O. Berger and Raphael¨ M. Jungers CPU Time for jordan for n n matrix with entries in {-128, ,127} Abstract— We study the algorithmic complexity of the prob- lem of deciding whether a Linear Time Invariant dynamical system with rational coefficients has bounded trajectories. Despite its ubiquitous and elementary nature in Systems and Control, it turns out that this question is quite intricate, and, to 100 the best of our knowledge, unsolved in the literature. We show that classical tools, such as Gaussian Elimination, the Routh– Hurwitz Criterion, and the Euclidean Algorithm for GCD of polynomials indeed allow for an algorithm that is polynomial 10-1 in the bit size of the instance. However, all these tools have to be implemented with care, and in a non-standard way, which relies on an advanced analysis. elapsed time [s] 10-2 I. INTRODUCTION This paper deals with the computational problem of de- ciding whether a Linear Time Invariant (LTI) dynamical system with rational coefficients has bounded trajectories; 1 2 3 4 5 6 7 8 9 10 11 12 see Problems 1 and 2 in Section II. We show that this n problem can be solved in polynomial time with respect to Fig. 1. A naive way to decide the boundedness of the trajectories of the bit size of the coefficients. We are interested in the exact a LTI system described by a rational matrix would be to use the Matlab complexity, also called “bit complexity” or “complexity in function jordan and look at the eigenvalues and size of the associated Jordan blocks. -

Permutation Matrices Whose Convex Combinations Are Orthostochastic
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Permutation Matrices Whose Convex Combinations Are Orthostochastic Yik-Hoi Au-Yeung and Che-Man Cheng Department of Mathematics University of Hong Kong Hong Kong Submitted by Richard A. Brualdi ABSTRACT Let P,, ., t’,,, be n x n permutation matrices. In this note, we give a simple necessary condition for all convex combinations of P,, . , Pm to be orthostochastic. We show that for n < 15 the condition is also sufficient, but for n > 15 whether the condition is suffkient or not is still open. 1. INTRODUCTION An n X n doubly stochastic (d.s.) matrix (aij> is said to be orthostochastic (o.s.) if there exists an n X n unitary matrix (a,,> such that aij = IuijJ2. OS. matrices play an important role in the consideration of generalized numerical ranges (for example, see [2]). For some other properties of OS. matrices see [4], [5], and [6]. For n > 3, it is known that there are d.s. matrices which are not 0.s. (for example, see [4]), and for n = 3 there are necessary and sufficient conditions for a d.s. matrix to be O.S. [2], but for n > 3 no such conditions have been obtained. We shall denote by II,, the group of all n x n permuta- tion matrices and by @n the set of all rr X n OS. matrices. Let P,, . , P,, E II,. We shall give a necessary condition for conv( P,, . , P,,,} c On, where “conv” means “convex hull of,” and then show that the same condition is also sufficient for n < I5. -

Perron–Frobenius Theorem - Wikipedia
Perron–Frobenius theorem - Wikipedia https://en.wikipedia.org/wiki/Perron–Frobenius_theorem Perron–Frobenius theorem In linear algebra, the Perron–Frobenius theorem, proved by Oskar Perron (1907) and Georg Frobenius (1912), asserts that a real square matrix with positive entries has a unique largest real eigenvalue and that the corresponding eigenvector can be chosen to have strictly positive components, and also asserts a similar statement for certain classes of nonnegative matrices. This theorem has important applications to probability theory (ergodicity of Markov chains); to the theory of dynamical systems (subshifts of finite type); to economics (Okishio's theorem,[1] Hawkins–Simon condition[2]); to demography (Leslie population age distribution model);[3] to social networks (DeGroot learning process), to Internet search engines[4] and even to ranking of football teams.[5] The first to discuss the ordering of players within tournaments using Perron– Frobenius eigenvectors is Edmund Landau.[6] [7] Contents Statement Positive matrices Non-negative matrices Classification of matrices Perron–Frobenius theorem for irreducible matrices Further properties Applications Non-negative matrices Stochastic matrices Algebraic graph theory Finite Markov chains Compact operators Proof methods Perron root is strictly maximal eigenvalue for positive (and primitive) matrices Proof for positive matrices Lemma Power method and the positive eigenpair Multiplicity one No other non-negative eigenvectors Collatz–Wielandt formula Perron projection as a limit: Ak/rk Inequalities for Perron–Frobenius eigenvalue Further proofs 1 of 18 2/8/19, 14:03 Perron–Frobenius theorem - Wikipedia https://en.wikipedia.org/wiki/Perron–Frobenius_theorem Perron projection Peripheral projection Cyclicity Caveats Terminology See also Notes References Original papers Further reading Statement Let positive and non-negative respectively describe matrices with exclusively positive real numbers as elements and matrices with exclusively non-negative real numbers as elements.