PDF of Y(T1, T2)Atx; Ϭ Not Done Here for the Sake of Simplicity and Generality
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

VMAA-Performance-Sta
Revised June 18, 2019 U.S. Department of Veterans Affairs (VA) Veteran Monthly Assistance Allowance for Disabled Veterans Training in Paralympic and Olympic Sports Program (VMAA) In partnership with the United States Olympic Committee and other Olympic and Paralympic entities within the United States, VA supports eligible service and non-service-connected military Veterans in their efforts to represent the USA at the Paralympic Games, Olympic Games and other international sport competitions. The VA Office of National Veterans Sports Programs & Special Events provides a monthly assistance allowance for disabled Veterans training in Paralympic sports, as well as certain disabled Veterans selected for or competing with the national Olympic Team, as authorized by 38 U.S.C. 322(d) and Section 703 of the Veterans’ Benefits Improvement Act of 2008. Through the program, VA will pay a monthly allowance to a Veteran with either a service-connected or non-service-connected disability if the Veteran meets the minimum military standards or higher (i.e. Emerging Athlete or National Team) in his or her respective Paralympic sport at a recognized competition. In addition to making the VMAA standard, an athlete must also be nationally or internationally classified by his or her respective Paralympic sport federation as eligible for Paralympic competition. VA will also pay a monthly allowance to a Veteran with a service-connected disability rated 30 percent or greater by VA who is selected for a national Olympic Team for any month in which the Veteran is competing in any event sanctioned by the National Governing Bodies of the Olympic Sport in the United State, in accordance with P.L. -

The USANA Compensation Plan (Malaysia)
The USANA Compensation Plan (Malaysia) Last revision: December, 2019 The USANA Compensation Plan encourages Figure 1. Distributors and Preferred Customers teamwork and ensures a fair distribution of income among Distributors, so you can build a stable leveraged income as your downline organisation grows. STARTING YOUR USANA BUSINESS YOU PreferredPC PreferredPC Customer Customer You start by joining as a member, your sponsor places you in an open position in his or her downline 1 3 4 2 organisation. As a USANA distributor, you may retail products to your friends, enroll them as Preferred Customers, or Distributor Distributor Distributor Distributor sponsor them into your organisation as Distributors (see Figure 1). In Malaysia, USANA allows you to build a maximum of four Distributor legs. AREAS OF INCOME: There are six ways to earn an income in USANA business: 3. WEEKLY COMMISSION (1) Retail Sales You earn weekly Commission based on the Group Sales (2) Front Line Commission Volume (GSV) of your global organisation. The GSV is the (3) Weekly Commission sum of all Sales Volume points from ALL the Distributors (4) Leadership Bonus and Preferred Customers in your organisation, irrespective (5) Elite Bonus of how many levels of referrals, and no matter where in the (6) Matching Bonus world they enroll. 1. RETAIL SALES The calculation for the Weekly Commission payout will be based on 20% of the total Group Sales Volume (GSV) You earn a retail profit by selling USANA products to your on the small side of the business.The minimum payout customers at the recommended retail prices which is 10% will be at 125 GSV and the maximum is 5000 GSV. -

United States Olympic Committee and U.S. Department of Veterans Affairs
SELECTION STANDARDS United States Olympic Committee and U.S. Department of Veterans Affairs Veteran Monthly Assistance Allowance Program The U.S. Olympic Committee supports Paralympic-eligible military veterans in their efforts to represent the USA at the Paralympic Games and other international sport competitions. Veterans who demonstrate exceptional sport skills and the commitment necessary to pursue elite-level competition are given guidance on securing the training, support, and coaching needed to qualify for Team USA and achieve their Paralympic dreams. Through a partnership between the United States Department of Veterans Affairs and the USOC, the VA National Veterans Sports Programs & Special Events Office provides a monthly assistance allowance for disabled Veterans of the Armed Forces training in a Paralympic sport, as authorized by 38 U.S.C. § 322(d) and section 703 of the Veterans’ Benefits Improvement Act of 2008. Through the program the VA will pay a monthly allowance to a Veteran with a service-connected or non-service-connected disability if the Veteran meets the minimum VA Monthly Assistance Allowance (VMAA) Standard in his/her respective sport and sport class at a recognized competition. Athletes must have established training and competition plans and are responsible for turning in monthly and/or quarterly forms and reports in order to continue receiving the monthly assistance allowance. Additionally, an athlete must be U.S. citizen OR permanent resident to be eligible. Lastly, in order to be eligible for the VMAA athletes must undergo either national or international classification evaluation (and be found Paralympic sport eligible) within six months of being placed on the allowance pay list. -

Para Cycling Information Sheet About the Sport Classification Explained
Para cycling information sheet About the sport Para cycling is cycling for people with impairments resulting from a health condition (disability). Para athletes with physical impairments either compete on handcycles, tricycles or bicycles, while those with a visual impairment compete on tandems with a sighted ‘pilot’. Para cycling is divided into track and road events, with seven events in total. Classification explained In Para sport classification provides the structure for fair and equitable competition to ensure that winning is determined by skill, fitness, power, endurance, tactical ability and mental focus – the same factors that account for success in sport for able-bodied athletes. The Para sport classification assessment process identifies the eligibility of each Para athlete’s impairment, and groups them into a sport class according to the degree of activity limitation resulting from their impairment. Classification is sport-specific as an eligible impairment affects a Para athlete’s ability to perform in different sports to a different extent. Each Para sport has a different classification system. Standard Classification in detail Para-Cycling sport classes include: Handcycle sport classes H1 – 5: There are five different sport classes for handcycle racing. The lower numbers indicate a more severe activity limitation. Para athletes competing in the H1 classes have a complete loss of trunk and leg function and limited arm function, e.g. as a result of a spinal cord injury. Para athletes in the H4 class have limited or no leg function, but good trunk and arm function. Para cyclists in sport classes H1 – 4 compete in a reclined position. Para cyclists in the H5 sport class sit on their knees because they are able to use their arms and trunk to accelerate the handcycle. -

Para Cycling
Cycling Canada Long-Term Development PARA-CYCLING i We acknowledge the financial support of the Government of Canada through Sport Canada, a branch of the Department of Canadian Heritage. CANADIAN CYCLING ASSOCIATION Long-Term Athlete Development: Para-cycling Contents 1 – Introduction . 2 2 – From Awareness to High Performance . 3 3 – Canada’s Para-cycling System: Overview and Para-cycling Stage by Stage . 10 4 – Building the Para-cycling System: Success Factors . 17 5 – Conclusion . 20 Resources and Contacts . 20 Acknowledgements Contributors: Mathieu Boucher, Stephen Burke, Julie Hutsebaut, Paul Jurbala, Sébastien Travers Assistance : Luc Arseneau, Louis Barbeau Credit : FQSC Para, No Accidental Champions, Photo from Rob Jones Document prepared by Paul Jurbala, communityactive Design: John Luimes, i . Design Group ISBN# 978-0-9809082-4-4 2013 CYCLING CANADA Long-Term Athlete Development: PARA-CYCLING 1 1 – Introduction Cycling Canada recognizes Long-Term Athlete Development (LTAD) as a corner- to providing LTAD-based programs from entry to Active for Life stages . Our stone for building cycling at all levels of competition and participation in Canada, goal is not simply to help Canadian Para-cyclists to be the best in the world, for athletes of all abilities . LTAD provides a progressive pathway for athletes to but to ensure that every athlete with an impairment can enjoy participation in optimize their development according to recognized stages and processes in cycling for a lifetime . human physical, mental, emotional, and cognitive maturation . LTAD is more than a model - it is a system and philosophy of sport development . The CCC’s Long-Term Athlete Development Guide presents a model for athlete LTAD is athlete-centered, coach-driven, and administration-supported . -

(VA) Veteran Monthly Assistance Allowance for Disabled Veterans
Revised May 23, 2019 U.S. Department of Veterans Affairs (VA) Veteran Monthly Assistance Allowance for Disabled Veterans Training in Paralympic and Olympic Sports Program (VMAA) In partnership with the United States Olympic Committee and other Olympic and Paralympic entities within the United States, VA supports eligible service and non-service-connected military Veterans in their efforts to represent the USA at the Paralympic Games, Olympic Games and other international sport competitions. The VA Office of National Veterans Sports Programs & Special Events provides a monthly assistance allowance for disabled Veterans training in Paralympic sports, as well as certain disabled Veterans selected for or competing with the national Olympic Team, as authorized by 38 U.S.C. 322(d) and Section 703 of the Veterans’ Benefits Improvement Act of 2008. Through the program, VA will pay a monthly allowance to a Veteran with either a service-connected or non-service-connected disability if the Veteran meets the minimum military standards or higher (i.e. Emerging Athlete or National Team) in his or her respective Paralympic sport at a recognized competition. In addition to making the VMAA standard, an athlete must also be nationally or internationally classified by his or her respective Paralympic sport federation as eligible for Paralympic competition. VA will also pay a monthly allowance to a Veteran with a service-connected disability rated 30 percent or greater by VA who is selected for a national Olympic Team for any month in which the Veteran is competing in any event sanctioned by the National Governing Bodies of the Olympic Sport in the United State, in accordance with P.L. -

Should the Para-Cycling Classification System Be Reclassified?
Should the para-cycling classification system be reclassified? David Borg1, John Osborne2, Johanna Liljedahl3, Michele Foster1, Carla Nooijen3 1The Hopkins Centre, Menzies Health Institute Queensland, Griffith University, Brisbane, Australia. 2School of Exercise and Nutrition Sciences, Queensland University of Technology, Brisbane, Australia. 3The Swedish School of Sport and Health Sciences (GIH), Stockholm, Sweden. Introduction Results Para-cycling athletes compete on bicycles (C1–5), handcycles The number of men in C1, C2, C3, C4 and C5 was 32, 76, 63, (H1–5) and tricycles (T1-2) in classes based on their functional 76, and 87, respectively. Nineteen men competed in T1 and 58 disability. Higher classes reflect less functional impairment. Para- in T2. The age range of men was 14–62 years. cycling classification is governed by the Union Cycliste Inter- The number of women competing in C1, C2, C3, C4 and C5 was nationale (UCI). The system aims to promote participation in the 4, 18, 16, 20 and 28, respectively. Eleven competed in T1 and sport, by controlling for the impact of impairment on the outcome 15 in T2. Women’s age ranged 17–55 years. of competition. Road race velocity for the bicycling and tricycling is shown in Recently, the separation between adjacent handcycling classes for Figure 1. Comparisons between adjacent classes for men and official UCI events was described––providing benchmarks for women are displayed in Table 2. active and potential athletes. Unfortunately, similar evaluation of the bicycling and tricycling disciplines has not been considered. With the expectation of C4 and C5 for women, the analysis showed that men's and women's road race performance was This study aimed to described the separation between adjacent statistically different between adjacent classes for bicycling and classes, based on performance, in UCI road race events for tricycling. -

Fire Department Fiscal Year 2020 Annual Report July 1, 2019 – June 30, 2020
Fire Department Fiscal Year 2020 Annual Report July 1, 2019 – June 30, 2020 Steven R. Goble Chief FIRE DEPARTMENT I. MISSION STATEMENT To preserve and protect life, property and the environment of Kaua‘i County from all hazards and emergencies. II. DEPARTMENT GOALS The Kaua‘i Fire Department (KFD) is dedicated and highly motivated to make our community safe. The KFD team has effectively carried out duties to achieve the department’s goals of preventing, suppressing and extinguishing all types of fires; responding to and mitigating any and all types of emergencies (medical, HAZMAT, search and rescue, disasters) in a highly trained, professional and safe manner; administering first aid and CPR to the basic life support level (EMT) and to make Kaua‘i a safer place by supporting and promoting training for the community; enforcing the National/Hawai‘i Fire Code and promote fire prevention through educational programs and community outreach; maintaining our vehicles and equipment for emergency response through a preventive maintenance program; and providing safe, guarded beaches through an effective and dynamic ocean safety program. III. PROGRAM DESCIPTION To accomplish these objectives, the department maintains a modern communications and record management system, conducts routine inspections, enforces and governs fire codes and regulations, conducts fire investigations to ascertain its origin and cause, promotes public awareness by providing on-going, up-to-date public education, advocates a structured and uniform fire training program which includes HAZMAT, CBRNE (chemical, biological, radiological, nuclear and explosive) and mountain and ocean search and rescue training, and promotes highly trained fire suppression and hazmat/rescue teams. -

SPECTATOR GUIDE U.S. Paralympics Cycling Open Cummings Research Park, Huntsville, Alabama April 17-18, 2021 Come Cheer for Our P
SPECTATOR GUIDE U.S. Paralympics Cycling Open Cummings Research Park, Huntsville, Alabama April 17-18, 2021 Come Cheer for our Paralympic Cyclists! The Huntsville/Madison County community is excited to host U.S. Paralympics Cycling on Saturday and Sunday, April 17-18, 2021 in Cummings Research Park. The U.S. Paralympics Cycling Open presented by Toyota, is one of four domestic cycling events and the second opportunity for Para-cyclists to qualify for the Summer Paralympics in Tokyo this summer. This is also the return to competitive racing for these outstanding athletes who have not competed in over a year due to the ongoing COVID-19 pandemic. In fact, this event has taken on more importance in the qualifying circuit as other international qualifying events have been cancelled due to the pandemic. We expect approximately 100 Para athletes to visit Huntsville. The athletes will compete in three different types of road cycling events including the men’s and women’s road race, individual time trial, and handcycling team relay. Learn more about U.S. Paralympics Cycling here: USParaCycling.org This guide serves as an FAQ. It provides information on how and where you can watch the action during race weekend. Link to: Parking Viewing Restrooms What’s on the Schedule Each Day Show Your Support! Athletes to Watch Page 1 Do I need tickets? No! The public is invited, and this is a free event. The events will happen rain or shine. Bring the family, pack a cooler, bring chairs and a blanket, and come watch along the outside ring of Explorer Boulevard, the loop of Cummings Research Park West. -
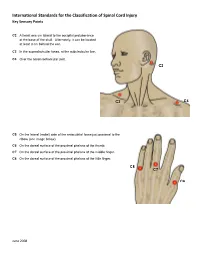
International Standards for the Classification of Spinal Cord Injury Key Sensory Points
International Standards for the Classification of Spinal Cord Injury Key Sensory Points C2 At least one cm lateral to the occipital protuberance at the base of the skull. Alternately, it can be located at least 3 cm behind the ear. C3 In the supraclavicular fossa, at the midclavicular line. C4 Over the acromioclavicular joint. C2 C3 C4 C5 On the lateral (radial) side of the antecubital fossa just proximal to the elbow (see image below). C6 On the dorsal surface of the proximal phalanx of the thumb. C7 On the dorsal surface of the proximal phalanx of the middle finger. C8 On the dorsal surface of the proximal phalanx of the little finger. C8 C7 C6 June 2008 International Standards for the Classification of Spinal Cord Injury Key Sensory Points C5 T2 T1 T1 On the medial (ulnar) side of the antecubital fossa, just proximal to the medial epicondyle of the humerus. T2 At the apex of the axilla. June 2008 International Standards for the Classification of Spinal Cord Injury Key Sensory Points T3 At the midclavicular line and the third intercostal space, found by palpating the anterior chest to locate the third C3 rib and the corresponding third intercostal space below it. T4 At the midclavicular line and the fourth intercostal space, located at the level of the nipples. T5 At the midclavicular line and the fifth intercostal space, located midway between the level of the nipples and T3 the level of the xiphisternum. T4 T6 At the midclavicular line, located at the level of the xiphisternum. T5 T7 At the midclavicular line, one quarter the distance between the level of T6 the xiphisternum and the level of the umbilicus. -

FDOT Context Classification Guide
FDOT Context Classification Guide Complete Streets Handbook The Florida Department of Transportation July 2020 Contents Chapter 1: FDOT Context Classification 4 INTRODUCTION TO CONTEXT CLASSIFICATION 7 DETERMINING PROJECT-SPECIFIC CONTEXT CLASSIFICATION 24 TRANSPORTATION CHARACTERISTICS Chapter 2: Linking Context Classification to the FDM and Other Documents 33 AASHTO’S A POLICY ON GEOMETRIC DESIGN OF HIGHWAYS AND STREETS 33 FDOT DESIGN MANUAL (FDM) 41 SPEED ZONING MANUAL 42 FDOT TRAFFIC ENGINEERING MANUAL 42 ACCESS MANAGEMENT 42 QUALITY/LEVEL OF SERVICE HANDBOOK 43 FLORIDA GREENBOOK Appendix A: Context Classification Case Studies Appendix B: Frequently Asked Questions Chapter 1: FDOT Context Classification It is the policy of the Florida Department of demand of the roadway is, and the challenges and Transportation (FDOT) to routinely plan, design, opportunities of each roadway user (see Figure construct, reconstruct, and operate a context- 1). The context classification and transportation sensitive system of streets in support of safety and characteristics of a roadway will determine key mobility. To this end, FDOT uses a context-based design criteria for all non-limited- access state approach to planning, designing, and operating the roadways. This Context Classification Guide provides state transportation network. FDOT has adopted a guidance on how context classification can be used, roadway classification system comprised of eight describes the measures used to determine the context classifications for all non-limited-access state -

Doping Control Guide for Testing Athletes in Para Sport
DOPING CONTROL GUIDE FOR TESTING ATHLETES IN PARA SPORT JULY 2021 INTERNATIONAL PARALYMPIC COMMITTEE 2 1 INTRODUCTION This guide is intended for athletes, anti-doping organisations and sample collection personnel who are responsible for managing the sample collection process – and other organisations or individuals who have an interest in doping control in Para sport. It provides advice on how to prepare for and manage the sample collection process when testing athletes who compete in Para sport. It also provides information about the Para sport classification system (including the types of impairments) and the types of modifications that may be required to complete the sample collection process. Appendix 1 details the classification system for those sports that are included in the Paralympic programme – and the applicable disciplines that apply within the doping control setting. The International Paralympic Committee’s (IPC’s) doping control guidelines outlined, align with Annex A Modifications for Athletes with Impairments of the World Anti-Doping Agency’s International Standard for Testing and Investigations (ISTI). It is recommended that anti-doping organisations (and sample collection personnel) follow these guidelines when conducting testing in Para sport. 2 DISABILITY & IMPAIRMENT In line with the United Nations Convention on the Rights of Persons with Disabilities (CRPD), ‘disability’ is a preferred word along with the usage of the term ‘impairment’, which refers to the classification system and the ten eligible impairments that are recognised in Para sports. The IPC uses the first-person language, i.e., addressing the athlete first and then their disability. As such, the right term encouraged by the IPC is ‘athlete or person with disability’.