1 Molecular Evidence for Sequential Colonization and Taxon Cycling In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Two Freshwater Shrimp Species of the Genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the Description of a New Species
A peer-reviewed open-access journal ZooKeys 923: 15–32 (2020) Caridina tetrazona 15 doi: 10.3897/zookeys.923.48593 RESEarcH articLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species Qing-Hua Chen1, Wen-Jian Chen2, Xiao-Zhuang Zheng2, Zhao-Liang Guo2 1 South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou 510520, Guangdong Province, China 2 Department of Animal Science, School of Life Science and Enginee- ring, Foshan University, Foshan 528231, Guangdong Province, China Corresponding author: Zhao-Liang Guo ([email protected]) Academic editor: I.S. Wehrtmann | Received 19 November 2019 | Accepted 7 February 2020 | Published 1 April 2020 http://zoobank.org/138A88CC-DF41-437A-BA1A-CB93E3E36D62 Citation: Chen Q-H, Chen W-J, Zheng X-Z, Guo Z-L (2020) Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species. ZooKeys 923: 15–32. https://doi.org/10.3897/zookeys.923.48593 Abstract A faunistic and ecological survey was conducted to document the diversity of freshwater atyid shrimps of Dawanshan Island. Two species of Caridina that occur on this island were documented and discussed. One of these, Caridina tetrazona sp. nov. is described and illustrated as new to science. It can be easily distinguished from its congeners based on a combination of characters, which includes a short rostrum, the shape of the endopod of the male first pleopod, the segmental ratios of antennular peduncle and third maxilliped, the slender scaphocerite, and the absence of a median projection on the posterior margin. -

New Zealand's Genetic Diversity
1.13 NEW ZEALAND’S GENETIC DIVERSITY NEW ZEALAND’S GENETIC DIVERSITY Dennis P. Gordon National Institute of Water and Atmospheric Research, Private Bag 14901, Kilbirnie, Wellington 6022, New Zealand ABSTRACT: The known genetic diversity represented by the New Zealand biota is reviewed and summarised, largely based on a recently published New Zealand inventory of biodiversity. All kingdoms and eukaryote phyla are covered, updated to refl ect the latest phylogenetic view of Eukaryota. The total known biota comprises a nominal 57 406 species (c. 48 640 described). Subtraction of the 4889 naturalised-alien species gives a biota of 52 517 native species. A minimum (the status of a number of the unnamed species is uncertain) of 27 380 (52%) of these species are endemic (cf. 26% for Fungi, 38% for all marine species, 46% for marine Animalia, 68% for all Animalia, 78% for vascular plants and 91% for terrestrial Animalia). In passing, examples are given both of the roles of the major taxa in providing ecosystem services and of the use of genetic resources in the New Zealand economy. Key words: Animalia, Chromista, freshwater, Fungi, genetic diversity, marine, New Zealand, Prokaryota, Protozoa, terrestrial. INTRODUCTION Article 10b of the CBD calls for signatories to ‘Adopt The original brief for this chapter was to review New Zealand’s measures relating to the use of biological resources [i.e. genetic genetic resources. The OECD defi nition of genetic resources resources] to avoid or minimize adverse impacts on biological is ‘genetic material of plants, animals or micro-organisms of diversity [e.g. genetic diversity]’ (my parentheses). -
W7192e19.Pdf
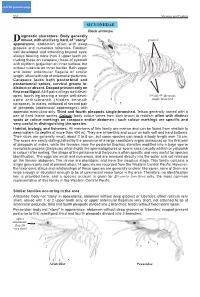
click for previous page 952 Shrimps and Prawns Sicyoniidae SICYONIIDAE Rock shrimps iagnostic characters: Body generally Drobust, with shell very hard, of “stony” grooves appearance; abdomen often with deep grooves and numerous tubercles. Rostrum well developed and extending beyond eyes, always bearing more than 3 upper teeth (in- cluding those on carapace); base of eyestalk with styliform projection on inner surface, but without tubercle on inner border. Both upper and lower antennular flagella of similar length, attached to tip of antennular peduncle. 1 Carapace lacks both postorbital and postantennal spines, cervical groove in- distinct or absent. Exopod present only on first maxilliped. All 5 pairs of legs well devel- 2 oped, fourth leg bearing a single well-devel- 3rd and 4th pleopods 4 single-branched oped arthrobranch (hidden beneath 3 carapace). In males, endopod of second pair 5 of pleopods (abdominal appendages) with appendix masculina only. Third and fourth pleopods single-branched. Telson generally armed with a pair of fixed lateral spines. Colour: body colour varies from dark brown to reddish; often with distinct spots or colour markings on carapace and/or abdomen - such colour markings are specific and very useful in distinguishing the species. Habitat, biology, and fisheries: All members of this family are marine and can be found from shallow to deep waters (to depths of more than 400 m). They are all benthic and occur on both soft and hard bottoms. Their sizes are generally small, about 2 to 8 cm, but some species can reach a body length over 15 cm. The sexes are easily distinguished by the presence of a large copulatory organ (petasma) on the first pair of pleopods of males, while the females have the posterior thoracic sternites modified into a large sperm receptacle process (thelycum) which holds the spermatophores or sperm sacs (usually whitish or yellowish in colour) after mating. -

Effects of Drought and Hurricane Disturbances on Headwater Distributions of Palaemonid River Shrimp (Macrobrachium Spp.) in the Luquillo Mountains, Puerto Rico
J. N. Am. Benthol. Soc., 2006, 25(1):99–107 Ó 2006 by The North American Benthological Society Effects of drought and hurricane disturbances on headwater distributions of palaemonid river shrimp (Macrobrachium spp.) in the Luquillo Mountains, Puerto Rico Alan P. Covich1 Institute of Ecology, University of Georgia, Athens, Georgia 30602-2202 USA 2 3 Todd A. Crowl AND Tamara Heartsill-Scalley Ecology Center and Department of Aquatic, Watershed, and Earth Sciences, Utah State University, Logan, Utah 84322 USA Abstract. Extreme events (hurricanes, floods, and droughts) can influence upstream migration of macroinvertebrates and wash out benthic communities, thereby locally altering food webs and species interactions. We sampled palaemonid river shrimp (Macrobrachium spp.), dominant consumers in headwaters of the Luquillo Mountains of northeastern Puerto Rico, to determine their distributions along an elevational gradient (274–456 m asl) during a series of disturbances (Hurricane Hugo in 1989, a drought in 1994, and Hurricane Georges in 1998) that occurred over a 15-y period (1988À2002). We measured shrimp abundance 3 to 6 times/y in Quebrada Prieta in the Espiritu Santo drainage as part of the Luquillo Long- Term Ecological Research Program. In general, Macrobrachium abundance declined with elevation during most years. The lowest mean abundance of Macrobrachium occurred during the 1994 drought, the driest year in 28 y of record in the Espiritu Santo drainage. Macrobrachium increased in abundance for 6 y following the 1994 drought. In contrast, hurricanes and storm flows had relatively little effect on Macrobrachium abundance. Key words: dispersal, drainage networks, geomorphology, habitat preference, omnivores, prey refugia. Flow-based events often modify the important roles (Covich et al. -

A Migratory Shrimp's Perspective on Habitat Fragmentation in The
A MIGRATORY SHRIMP’S PERSPECTIVE ON HABITAT FRAGMENTATION IN THE NEOTROPICS: EXTENDING OUR KNOWLEDGE FROM PUERTO RICO BY MARCIA N. SNYDER1,3), ELIZABETH P. ANDERSON2,4) and CATHERINE M. PRINGLE1,5) 1) Odum School of Ecology, University of Georgia, Athens, Georgia 30602, U.S.A. 2) Global Water for Sustainability Program, Florida International University, Miami, FL 33199, U.S.A. ABSTRACT Migratory freshwater fauna depend on longitudinal connectivity of rivers throughout their life cycles. Amphidromous shrimps spend their adult life in freshwater but their larvae develop into juveniles in salt water. River fragmentation resulting from pollution, land use change, damming and water withdrawals can impede dispersal and colonization of larval shrimps. Here we review current knowledge of river fragmentation effects on freshwater amphidromous shrimp in the Neotropics, with a focus on Puerto Rico and Costa Rica. In Puerto Rico, many studies have contributed to our knowledge of the natural history and ecological role of migratory neotropical shrimps, whereas in Costa Rica, studies of freshwater migratory shrimp have just begun. Here we examine research findings from Puerto Rico and the applicability of those findings to continental Costa Rica. Puerto Rico has a relatively large number of existing dams and water withdrawals, which have heavily fragmented rivers. The effects of fragmentation on migratory shrimps’ distribution have been documented on the landscape-scale in Puerto Rico. Over the last decade, dams for hydropower production have been constructed on rivers throughout Costa Rica. In both countries, large dams restrict shrimps from riverine habitat in central highland regions; in Puerto Rico 27% of stream kilometers are upstream of large dams while in Costa Rica 10% of stream kilometers are upstream of dams. -

Los Camarones Del Bosque Nacional El Yunque
i te fijas bien en los ríos y las pozas Pueden nadar rápidamente hacia atrás por cortos Y...¿Cómo se reproducen los camarones ? de El Yunque puedes que veas algo periódos de tiempo haciendo un rápido moverse rápidamente por el fondo. movimiento o flexión en el abdomen. con el rabo. La reproducción y etapas de la vida de un Lo más probable hayas visto una de camarón son muy inetersantes. La época Slas 10 especies de camarones que habitan en Las etapas de la vida de un camarón son muy reproductiva de los camarones ocurre durante las aguas del interesantes. El camarón adulto vive en charcas y los meses de verano aunque hay especies que se bosque. La mayoría rápidos y por la noche se concentran sobre reproducen durante todo el año. de los sistemas de piedras, ramas de árboles y hojas pues es un agua dulce y animal de hábitos nocturnos. El apareamiento se lleva a cabo cuando el salobre de Puerto macho se coloca en ángulo recto con la hembra Rico están Micratya poeyi Especies y Hábitos alimenticios y le transfiere un espermatóforo a un habitados por receptáculo en el abdomen de la hembra. De 6 a camarones de las familias Atyidae y En Puerto Rico existen 9 especies de la familia 20 horas después del apareamiento, la hembra Palaemonidae. Atyidae y de éstas seis 6 se encuentran en los ríos carga bajo su abdomen una gran cantidad de de El Yunque. huevos. Esta cantidad depende de la especie y Estas curiosas especies de animales tienen Los miembros de el individuo. -

Molecular Evidence for Sequential Colonization and Taxon Cycling In
Molecular Ecology (2008) 17, 1066–1075 doi: 10.1111/j.1365-294X.2007.03637.x MolecularBlackwell Publishing Ltd evidence for sequential colonization and taxon cycling in freshwater decapod shrimps on a Caribbean island BENJAMIN D. COOK,* CATHERINE M. PRINGLE† and JANE M. HUGHES* *Australian Rivers Institute, Griffith University, Nathan, Queensland, Australia, †Institute of Ecology, University of Georgia, Athens, GA, USA Abstract Taxon cycling, i.e. sequential phases of expansions and contractions in species’ distributions associated with ecological or morphological shifts, are postulated to characterize dynamic biogeographic histories in various island faunas. The Caribbean freshwater shrimp assemblage is mostly widespread and sympatric throughout the region, although one species (Atyidae: Atya lanipes) is geographically restricted and ecologically and morphologically differentiated from other Atya species. Using patterns of nucleotide variation at the COI mtDNA gene in five species of freshwater shrimp (A. lanipes, A. scabra, A. innocuous; Xiphocarididae: Xiphocaris elongata; Palaemonidae: Macrobrachium faustinum) from Puerto Rico, we expected to detect a signature of sequential colonization in these shrimp, consistent with the concept of taxon cycling, and expected that A. lanipes would be at a different taxon stage (i.e. an early stage species) to all other species. We also examined patterns of genetic population structure in each species expected with poor, intermediate and well-developed abilities for among-river dispersal. Population expansions were detected in all species, although the relative timing of the expansions varied among them. Assuming that population expansions followed colonization of Puerto Rico by freshwater shrimp, results bear the hallmarks of sequential colonization and taxon cycling in this fauna. A. -

Perspectives on Typhlatya (Crustacea, Decapoda)
Contributions to Zoology, 65 (2) 79-99 (1995) SPB Academic Publishing bv, Amsterdam New perspectives on the evolution of the genus Typhlatya (Crustacea, Decapoda): first record of a cavernicolous atyid in the Iberian Peninsula, Typhlatya miravetensis n. sp. Sebastián Sanz & Dirk Platvoet 1 Unitat d'Ecologia, Facultat de Ciències Biologiques, Universitat de Valencia, E-46100 Burjassot, 2 Valencia, Spain; Institutefor Systematics and Population Biology (Zoological Museum, Amsterdam), University of Amsterdam, P.O. Box 94766, 1090 GT Amsterdam, The Netherlands Keywords: Typhlatya, Decapoda, Spain, subterranean waters, systematics, zoogeography, vicariance, evolution, key to genus Abstract historia geológica de la zona y la distribución mundial del género, del grupo de géneros, y la familia. On several occasions, shrimps belonging to a new species ofthe genus Typhlatya were collected in a cave in the province of Castellón, Spain. This is the first record of the in the genus Introduction Iberian Peninsula. The species is described and the validity, dis- tribution, and zoogeography of the genus, as well as the status In 1993 and were on several of the discussed. 1994, shrimps caught genus Spelaeocaris, are Former models for the occasions in in the evolution of the genus Typhlatya and its genus group are re- a cave near Cabanes, province viewed, as well asthe system ofinner classification of the Atyidae of Castellón, eastern Spain. The specimens belong and its For the and evolution of biogeographical meaning. age the to genus Typhlatya Creaser, 1936, a genus the genus we developed a new model based on vicariance prin- with members known from the Galápagos Islands, ciples that involves further evolution of each species after the Ascension and the Caribbean of the ancestral This allows estimations Island, Bermuda, disruption range. -

Behavioral Responses of the Endemic Shrimp Halocaridina Rubra
Behavioral Responses of the Endemic Shrimp Halocaridina rubra (Malacostraca: Atyidae) to an Introduced Fish, Gambusia affinis (Actinopterygii: Poeciliidae) and Implications for the Trophic Structure of Hawaiian Anchialine Ponds1 Krista A. Capps,2 Caroline B. Turner,2 Michael T. Booth,2 Danica L. Lombardozzi,2 Scott H. McArt,4 David Chai,3 and Nelson G. Hairston Jr.2,5 Abstract: In the Hawaiian Islands, intentionally introduced exotic fishes have been linked to changes in native biodiversity and community composition. In 1905, the mosquito fish Gambusia affinis was introduced to control mosquitoes. Subsequently, G. affinis spread throughout the Islands and into coastal anchia- line ponds. Previous studies suggest that presence of invasive fishes in anchialine ponds may eliminate native species, including the endemic shrimp Halocaridina rubra. We examined effects of G. affinis on H. rubra populations in anchialine ponds on the Kona-Kohala coast of the island of Hawai‘i. In the presence of G. affinis, H. rubra exhibited a diel activity pattern that was not seen in fishless ponds. Shrimp in ponds with fish were active only at night. This pattern was ev- ident in anchialine ponds and in laboratory experiments. In laboratory predation experiments, G. affinis preferentially consumed smaller H. rubra, and in the field the H. rubra collected from invaded sites were larger than those from fishless ponds. Analysis of trophic position using stable isotope analyses showed that feeding of H. rubra was not significantly distinct from that of snails, assumed to feed at trophic level 2.0 on epilithic algae, but G. affinis was slightly omnivo- rous, feeding at tropic level 2.2. -

Cave Biodiversity of the Southern Cumberland Plateau Kirk S
b-3-guidebook_Guidebook3 6/18/2014 10:01 PM Page 159 Cave Biodiversity of the Southern Cumberland Plateau Kirk S. Zigler, NSS 62696; Matthew L. Niemiller, NSS 53235; and Danté B. Fenolio The South Cumberland Region of Tennessee, Alabama, and Georgia (Figure 1) is known for its tremendous diversity of caves, including huge pits, massive stream passages, and tight crawls. Less well known is that the region also supports tremendous cave biodiversity (Niemiller, Zigler, and Fenolio, 2013). Here we discuss many of the species that inhabit caves of the region, focusing on the southern Cumberland Plateau. Cave Biodiversity Four ecological classes of organisms can be found in caves: trogloxenes, subtroglophiles, eutroglophiles, and troglobionts (Culver and Pipan, 2009). Trogloxenes are not typically found in caves and cannot persist there for long periods of time. They must either find their way back to the surface or ultimately perish. Subtroglophiles are commonly found in caves but are associated with surface habitats for at least part of their life cycle. Some are seasonal inhabitants of caves and others move back and forth from cave to surface habitats for feeding, such as cave-roosting bats, cave crickets, and Allegheny Woodrats (Neotoma magister). Eutroglophiles are commonly found underground but can be found in surface habitats. Unlike trogloxenes and subtroglophiles, eutroglophiles can complete their entire life cycle Figure 1 - The South Cumberland Region at the junction of underground. Examples include the Cave Salamander Tennessee, Alabama, and Georgia. Figure courtesy of Nick Hollingshead. (Eurycea lucifuga) and the Cave Orbweaver (Meta ovalis). Troglobionts are obligate, permanent residents of subterranean habitats. -

Annotated Checklist of New Zealand Decapoda (Arthropoda: Crustacea)
Tuhinga 22: 171–272 Copyright © Museum of New Zealand Te Papa Tongarewa (2011) Annotated checklist of New Zealand Decapoda (Arthropoda: Crustacea) John C. Yaldwyn† and W. Richard Webber* † Research Associate, Museum of New Zealand Te Papa Tongarewa. Deceased October 2005 * Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, New Zealand ([email protected]) (Manuscript completed for publication by second author) ABSTRACT: A checklist of the Recent Decapoda (shrimps, prawns, lobsters, crayfish and crabs) of the New Zealand region is given. It includes 488 named species in 90 families, with 153 (31%) of the species considered endemic. References to New Zealand records and other significant references are given for all species previously recorded from New Zealand. The location of New Zealand material is given for a number of species first recorded in the New Zealand Inventory of Biodiversity but with no further data. Information on geographical distribution, habitat range and, in some cases, depth range and colour are given for each species. KEYWORDS: Decapoda, New Zealand, checklist, annotated checklist, shrimp, prawn, lobster, crab. Contents Introduction Methods Checklist of New Zealand Decapoda Suborder DENDROBRANCHIATA Bate, 1888 ..................................... 178 Superfamily PENAEOIDEA Rafinesque, 1815.............................. 178 Family ARISTEIDAE Wood-Mason & Alcock, 1891..................... 178 Family BENTHESICYMIDAE Wood-Mason & Alcock, 1891 .......... 180 Family PENAEIDAE Rafinesque, 1815 .................................. -

Universidade De São Paulo Ffclrp
UNIVERSIDADE DE SÃO PAULO FFCLRP - DEPARTAMENTO DE BIOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA COMPARADA Avaliação sistemática de camarões de água doce do gênero Atya Leach, 1816 (Crustacea: Decapoda: Atyidae) por meio de dados moleculares Caio Martins Cruz Alves de Oliveira Dissertação apresentada à Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto da USP, como parte das exigências para a obtenção do título de Mestre em Ciências, Área: BIOLOGIA COMPARADA Ribeirão Preto - SP 2017 UNIVERSIDADE DE SÃO PAULO FFCLRP - DEPARTAMENTO DE BIOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA COMPARADA Avaliação sistemática de camarões de água doce do gênero Atya Leach, 1816 (Crustacea: Decapoda: Atyidae) por meio de dados moleculares Caio Martins Cruz Alves de Oliveira Orientador: Prof. Dr. Fernando Luis Medina Mantelatto Co-orientadora: Profa. Dra. Mariana Terossi Rodrigues Mariano Dissertação apresentada à Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto da USP, como parte das exigências para a obtenção do título de Mestre em Ciências, Área: BIOLOGIA COMPARADA Versão Original Ribeirão Preto - SP 2017 Autorizo a reprodução e divulgação total ou parcial deste trabalho, por qualquer meio convencional ou eletrônico, para fins de estudo e pesquisa, desde que citada a fonte. Oliveira, C. M. C. A. “Avaliação sistemática de camarões de água doce do gênero Atya Leach, 1816 (Crustacea: Decapoda: Atyidae) por meio de dados moleculares” Ribeirão Preto, 2017 vii+107p. Dissertação (Mestrado – Programa de Pós-graduação em Ciências. Área de concentração: Biologia Comparada). Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Universidade de São Paulo (FFCLRP-USP). Orientador: Mantelatto, F.L.M.; Co-orientadora: Mariano, M.T.R.