K.Gobalan Et.Al APPLICATIONS of BIOINFORMATICS in GENOMICS
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bioinformatics 1
Bioinformatics 1 Bioinformatics School School of Science, Engineering and Technology (http://www.stmarytx.edu/set/) School Dean Ian P. Martines, Ph.D. ([email protected]) Department Biological Science (https://www.stmarytx.edu/academics/set/undergraduate/biological-sciences/) Bioinformatics is an interdisciplinary and growing field in science for solving biological, biomedical and biochemical problems with the help of computer science, mathematics and information technology. Bioinformaticians are in high demand not only in research, but also in academia because few people have the education and skills to fill available positions. The Bioinformatics program at St. Mary’s University prepares students for graduate school, medical school or entry into the field. Bioinformatics is highly applicable to all branches of life sciences and also to fields like personalized medicine and pharmacogenomics — the study of how genes affect a person’s response to drugs. The Bachelor of Science in Bioinformatics offers three tracks that students can choose. • Bachelor of Science in Bioinformatics with a minor in Biology: 120 credit hours • Bachelor of Science in Bioinformatics with a minor in Computer Science: 120 credit hours • Bachelor of Science in Bioinformatics with a minor in Applied Mathematics: 120 credit hours Students will take 23 credit hours of core Bioinformatics classes, which included three credit hours of internship or research and three credit hours of a Bioinformatics Capstone course. BS Bioinformatics Tracks • Bachelor of Science -

13 Genomics and Bioinformatics
Enderle / Introduction to Biomedical Engineering 2nd ed. Final Proof 5.2.2005 11:58am page 799 13 GENOMICS AND BIOINFORMATICS Spencer Muse, PhD Chapter Contents 13.1 Introduction 13.1.1 The Central Dogma: DNA to RNA to Protein 13.2 Core Laboratory Technologies 13.2.1 Gene Sequencing 13.2.2 Whole Genome Sequencing 13.2.3 Gene Expression 13.2.4 Polymorphisms 13.3 Core Bioinformatics Technologies 13.3.1 Genomics Databases 13.3.2 Sequence Alignment 13.3.3 Database Searching 13.3.4 Hidden Markov Models 13.3.5 Gene Prediction 13.3.6 Functional Annotation 13.3.7 Identifying Differentially Expressed Genes 13.3.8 Clustering Genes with Shared Expression Patterns 13.4 Conclusion Exercises Suggested Reading At the conclusion of this chapter, the reader will be able to: & Discuss the basic principles of molecular biology regarding genome science. & Describe the major types of data involved in genome projects, including technologies for collecting them. 799 Enderle / Introduction to Biomedical Engineering 2nd ed. Final Proof 5.2.2005 11:58am page 800 800 CHAPTER 13 GENOMICS AND BIOINFORMATICS & Describe practical applications and uses of genomic data. & Understand the major topics in the field of bioinformatics and DNA sequence analysis. & Use key bioinformatics databases and web resources. 13.1 INTRODUCTION In April 2003, sequencing of all three billion nucleotides in the human genome was declared complete. This landmark of modern science brought with it high hopes for the understanding and treatment of human genetic disorders. There is plenty of evidence to suggest that the hopes will become reality—1631 human genetic diseases are now associated with known DNA sequences, compared to the less than 100 that were known at the initiation of the Human Genome Project (HGP) in 1990. -

Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India Meera Yadav University of Delhi, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Library Philosophy and Practice (e-journal) Libraries at University of Nebraska-Lincoln 2015 Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India meera yadav University of Delhi, [email protected] Manlunching Tawmbing Saha Institute of Nuclear Physisc, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/libphilprac Part of the Library and Information Science Commons yadav, meera and Tawmbing, Manlunching, "Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India" (2015). Library Philosophy and Practice (e-journal). 1254. http://digitalcommons.unl.edu/libphilprac/1254 Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India Dr Meera, Manlunching Department of Library and Information Science, University of Delhi, India [email protected], [email protected] Abstract Information plays a vital role in Bioinformatics to achieve the existing Bioinformatics information technologies. Librarians have to identify the information needs, uses and problems faced to meet the needs and requirements of the Bioinformatics users. The paper analyses the response of 315 Bioinformatics users of 15 Bioinformatics centers in India. The papers analyze the data with respect to use of different Bioinformatics databases and tools used by scholars and scientists, areas of major research in Bioinformatics, Major research project, thrust areas of research and use of different resources by the user. The study identifies the various Bioinformatics services and resources used by the Bioinformatics researchers. Keywords: Informaion services, Users, Inforamtion needs, Bioinformatics resources 1. Introduction ‘Needs’ refer to lack of self-sufficiency and also represent gaps in the present knowledge of the users. -

Challenges in Bioinformatics
Yuri Quintana, PhD, delivered a webinar in AllerGen’s Webinars for Research Success series on February 27, 2018, discussing different approaches to biomedical informatics and innovations in big-data platforms for biomedical research. His main messages and a hyperlinked index to his presentation follow. WHAT IS BIOINFORMATICS? Bioinformatics is an interdisciplinary field that develops analytical methods and software tools for understanding clinical and biological data. It combines elements from many fields, including basic sciences, biology, computer science, mathematics and engineering, among others. WHY DO WE NEED BIOINFORMATICS? Chronic diseases are rapidly expanding all over the world, and associated healthcare costs are increasing at an astronomical rate. We need to develop personalized treatments tailored to the genetics of increasingly diverse patient populations, to clinical and family histories, and to environmental factors. This requires collecting vast amounts of data, integrating it, and making it accessible and usable. CHALLENGES IN BIOINFORMATICS Data collection, coordination and archiving: These technologies have evolved to the point Many sources of biomedical data—hospitals, that we can now analyze not only at the DNA research centres and universities—do not have level, but down to the level of proteins. DNA complete data for any particular disease, due in sequencers, DNA microarrays, and mass part to patient numbers, but also to the difficulties spectrometers are generating tremendous of data collection, inter-operability -

Comparing Bioinformatics Software Development by Computer Scientists and Biologists: an Exploratory Study
Comparing Bioinformatics Software Development by Computer Scientists and Biologists: An Exploratory Study Parmit K. Chilana Carole L. Palmer Amy J. Ko The Information School Graduate School of Library The Information School DUB Group and Information Science DUB Group University of Washington University of Illinois at University of Washington [email protected] Urbana-Champaign [email protected] [email protected] Abstract Although research in bioinformatics has soared in the last decade, much of the focus has been on high- We present the results of a study designed to better performance computing, such as optimizing algorithms understand information-seeking activities in and large-scale data storage techniques. Only recently bioinformatics software development by computer have studies on end-user programming [5, 8] and scientists and biologists. We collected data through semi- information activities in bioinformatics [1, 6] started to structured interviews with eight participants from four emerge. Despite these efforts, there still is a gap in our different bioinformatics labs in North America. The understanding of problems in bioinformatics software research focus within these labs ranged from development, how domain knowledge among MBB computational biology to applied molecular biology and and CS experts is exchanged, and how the software biochemistry. The findings indicate that colleagues play a development process in this domain can be improved. significant role in information seeking activities, but there In our exploratory study, we used an information is need for better methods of capturing and retaining use perspective to begin understanding issues in information from them during development. Also, in bioinformatics software development. We conducted terms of online information sources, there is need for in-depth interviews with 8 participants working in 4 more centralization, improved access and organization of bioinformatics labs in North America. -

Database Technology for Bioinformatics from Information Retrieval to Knowledge Systems
Database Technology for Bioinformatics From Information Retrieval to Knowledge Systems Luis M. Rocha Complex Systems Modeling CCS3 - Modeling, Algorithms, and Informatics Los Alamos National Laboratory, MS B256 Los Alamos, NM 87545 [email protected] or [email protected] 1 Molecular Biology Databases 3 Bibliographic databases On-line journals and bibliographic citations – MEDLINE (1971, www.nlm.nih.gov) 3 Factual databases Repositories of Experimental data associated with published articles and that can be used for computerized analysis – Nucleic acid sequences: GenBank (1982, www.ncbi.nlm.nih.gov), EMBL (1982, www.ebi.ac.uk), DDBJ (1984, www.ddbj.nig.ac.jp) – Amino acid sequences: PIR (1968, www-nbrf.georgetown.edu), PRF (1979, www.prf.op.jp), SWISS-PROT (1986, www.expasy.ch) – 3D molecular structure: PDB (1971, www.rcsb.org), CSD (1965, www.ccdc.cam.ac.uk) Lack standardization of data contents 3 Knowledge Bases Intended for automatic inference rather than simple retrieval – Motif libraries: PROSITE (1988, www.expasy.ch/sprot/prosite.html) – Molecular Classifications: SCOP (1994, www.mrc-lmb.cam.ac.uk) – Biochemical Pathways: KEGG (1995, www.genome.ad.jp/kegg) Difference between knowledge and data (semiosis and syntax)?? 2 Growth of sequence and 3D Structure databases Number of Entries 3 Database Technology and Bioinformatics 3 Databases Computerized collection of data for Information Retrieval Shared by many users Stored records are organized with a predefined set of data items (attributes) Managed by a computer program: the database -
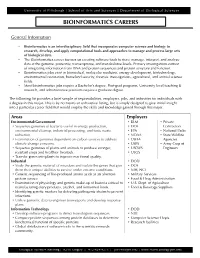
Bioinformatics Careers
University of Pittsburgh | School of Arts and Sciences | Department of Biological Sciences BIOINFORMATICS CAREERS General Information • Bioinformatics is an interdisciplinary field that incorporates computer science and biology to research, develop, and apply computational tools and approaches to manage and process large sets of biological data. • The Bioinformatics career focuses on creating software tools to store, manage, interpret, and analyze data at the genome, proteome, transcriptome, and metabalome levels. Primary investigations consist of integrating information from DNA and protein sequences and protein structure and function. • Bioinformatics jobs exist in biomedical, molecular medicine, energy development, biotechnology, environmental restoration, homeland security, forensic investigations, agricultural, and animal science fields. • Most bioinformatics jobs require a Bachelor’s degree. Post-grad programs, University level teaching & research, and administrative positions require a graduate degree. The following list provides a brief sample of responsibilities, employers, jobs, and industries for individuals with a degree in this major. This is by no means an exhaustive listing, but is simply designed to give initial insight into a particular career field that would employ the skills and knowledge gained through this major. Areas Employers Environmental/Government • BLM • Private • Sequence genomes of bacteria useful in energy production, • DOE Contractors environmental cleanup, industrial processing, and toxic waste • EPA • National Parks reduction. • NOAA • State Wildlife • Examination of genomes dependent on carbon sources to address • USDA Agencies climate change concerns. • USFS • Army Corp of • Sequence genomes of plants and animals to produce stronger, • USFWS Engineers resistant crops and healthier livestock. • USGS • Transfer genes into plants to improve nutritional quality. Industrial • DOD • Study the genetic material of microbes and isolate the genes that give • DOE them their unique abilities to survive under extreme conditions. -

Problems for Structure Learning: Aggregation and Computational Complexity
(in press). In S. Das, D. Caragea, W. H. Hsu, & S. M. Welch (Eds.), Computational methodologies in gene regulatory networks. Hershey, PA: IGI Global Publishing. Problems for Structure Learning: Aggregation and Computational Complexity Frank Wimberly Carnegie Mellon University (retired), USA David Danks Carnegie Mellon University and Institute for Human & Machine Cognition, USA Clark Glymour Carnegie Mellon University and Institute for Human & Machine Cognition, USA Tianjiao Chu University of Pittsburgh, USA ABSTRACT Machine learning methods to find graphical models of genetic regulatory networks from cDNA microarray data have become increasingly popular in recent years. We provide three reasons to question the reliability of such methods: (1) a major theoretical challenge to any method using conditional independence relations; (2) a simulation study using realistic data that confirms the importance of the theoretical challenge; and (3) an analysis of the computational complexity of algorithms that avoid this theoretical challenge. We have no proof that one cannot possibly learn the structure of a genetic regulatory network from microarray data alone, nor do we think that such a proof is likely. However, the combination of (i) fundamental challenges from theory, (ii) practical evidence that those challenges arise in realistic data, and (iii) the difficulty of avoiding those challenges leads us to conclude that it is unlikely that current microarray technology will ever be successfully applied to this structure learning problem. INTRODUCTION An important goal of cell biology is to understand the network of dependencies through which genes in a tissue type regulate the synthesis and concentrations of protein species. A mediating step in such synthesis is the production of messenger RNA (mRNA). -

Spontaneous Generation & Origin of Life Concepts from Antiquity to The
SIMB News News magazine of the Society for Industrial Microbiology and Biotechnology April/May/June 2019 V.69 N.2 • www.simbhq.org Spontaneous Generation & Origin of Life Concepts from Antiquity to the Present :ŽƵƌŶĂůŽĨ/ŶĚƵƐƚƌŝĂůDŝĐƌŽďŝŽůŽŐLJΘŝŽƚĞĐŚŶŽůŽŐLJ Impact Factor 3.103 The Journal of Industrial Microbiology and Biotechnology is an international journal which publishes papers in metabolic engineering & synthetic biology; biocatalysis; fermentation & cell culture; natural products discovery & biosynthesis; bioenergy/biofuels/biochemicals; environmental microbiology; biotechnology methods; applied genomics & systems biotechnology; and food biotechnology & probiotics Editor-in-Chief Ramon Gonzalez, University of South Florida, Tampa FL, USA Editors Special Issue ^LJŶƚŚĞƚŝĐŝŽůŽŐLJ; July 2018 S. Bagley, Michigan Tech, Houghton, MI, USA R. H. Baltz, CognoGen Biotech. Consult., Sarasota, FL, USA Impact Factor 3.500 T. W. Jeffries, University of Wisconsin, Madison, WI, USA 3.000 T. D. Leathers, USDA ARS, Peoria, IL, USA 2.500 M. J. López López, University of Almeria, Almeria, Spain C. D. Maranas, Pennsylvania State Univ., Univ. Park, PA, USA 2.000 2.505 2.439 2.745 2.810 3.103 S. Park, UNIST, Ulsan, Korea 1.500 J. L. Revuelta, University of Salamanca, Salamanca, Spain 1.000 B. Shen, Scripps Research Institute, Jupiter, FL, USA 500 D. K. Solaiman, USDA ARS, Wyndmoor, PA, USA Y. Tang, University of California, Los Angeles, CA, USA E. J. Vandamme, Ghent University, Ghent, Belgium H. Zhao, University of Illinois, Urbana, IL, USA 10 Most Cited Articles Published in 2016 (Data from Web of Science: October 15, 2018) Senior Author(s) Title Citations L. Katz, R. Baltz Natural product discovery: past, present, and future 103 Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and R. -

B.Sc. in Bioinformatics (Three Years)
MAULANA ABUL KALAM AZAD UNIVERSITY OF TECHNOLOGY, WEST BENGAL NH-12 (Old NH-34), Simhat, Haringhata, Nadia -741249 Department of Bioinformatics B.Sc. in Bioinformatics Choice Based Credit System 144 Credits for 3-Year UG MAKAUT Framework w.e.f. AY 2020-21 MODEL CURRICULUM For B.Sc. in Bioinformatics MAULANA ABUL KALAM AZAD UNIVERSITY OF TECHNOLOGY, WEST BENGAL NH-12 (Old NH-34), Simhat, Haringhata, Nadia -741249 Department of Bioinformatics B.Sc. in Bioinformatics Course Title: Course Name: B.Sc. in Bioinformatics Formal Abbreviation: BSBIN Organizational Arrangements: Managing Faculty: Faculty from the Department of Bioinformatics Collaborating Faculties: Professionals from the Department of Biotechnology, Mathematics, English, Chemistry, Physics etc. External Partners: To be decided Nature of Development: - a new course. Objective: B.Sc.in Bioinformatics (BSBIN) is one of the most sought after career oriented professional programs offered at the bachelor’s level. This degree course opens up innumerable career options and opportunities to the aspiring bioinformatics professionals both in India and abroad. This program offers to gain knowledge about multidisciplinary science with triple major combination of biology, mathematics and computer science. The course is MAULANA ABUL KALAM AZAD UNIVERSITY OF TECHNOLOGY, WEST BENGAL NH-12 (Old NH-34), Simhat, Haringhata, Nadia -741249 Department of Bioinformatics B.Sc. in Bioinformatics closely attached with analytical and numerical methods of applied biological, mathematical and computational problems to address and solve pressing challenges in all areas of science and engineering. Subjects are taught in this program with hands on experience in Bioinformatics, drug discovery, genomics and proteomics, Molecular Biology and Genetics, Microbiology and Biochemsitry, Computer programming, Biostatistics, Mathematical Modelling etc. -

Bioinformatics/Computational Biology/Data Science
Bioinformatics/Computational biology/Data Science Potential Careers (includes, but not limited to): Academic Independent research: Running an independent research program at a University. Requires a PhD in Biology or related field, with significant training in Bioinformatics/Computational Biology. Lab may specialize in computational biology or use it as a tool to support wet-lab studies. Research Assistant: Working within a laboratory for independent researcher(s), using computational skills to help solve biological problems. MS would be helpful, but not required. Research support staff: Working in an institutional support facility, helping researchers to find computational solutions to their problems. MS or PhD would be helpful, but not required, depending on the position. Software developer – No formal education required. Healthcare informatics: Working for a hospital or healthcare system to gather data and analyze it, often supporting health care missions to promote best practices. E.g. a surgical team would like to present data that their procedure is superior to others. They ask you to gather data from surveys and medical records on the outcomes (mortality, infection rates, recovery time, patient satisfaction, ect), analyze it and summarize the outcome. Each of these careers may be regular salary/hourly employment or contract employment where you are hired to perform a specific task for a fee. Genetic Counseling – M.S. Required Examples of Grad programs (not a complete list) o IUPUI – 1 yr MS program in applied data science or Health informatics. Offer “Bootcamp” (extra semester of training in computational aspects). Also offer multiple online courses in computer coding. o NYU – MS is Biomedical Informatics Training for CMB students: B.S. -

Algorithmic Complexity in Computational Biology: Basics, Challenges and Limitations
Algorithmic complexity in computational biology: basics, challenges and limitations Davide Cirillo#,1,*, Miguel Ponce-de-Leon#,1, Alfonso Valencia1,2 1 Barcelona Supercomputing Center (BSC), C/ Jordi Girona 29, 08034, Barcelona, Spain 2 ICREA, Pg. Lluís Companys 23, 08010, Barcelona, Spain. # Contributed equally * corresponding author: [email protected] (Tel: +34 934137971) Key points: ● Computational biologists and bioinformaticians are challenged with a range of complex algorithmic problems. ● The significance and implications of the complexity of the algorithms commonly used in computational biology is not always well understood by users and developers. ● A better understanding of complexity in computational biology algorithms can help in the implementation of efficient solutions, as well as in the design of the concomitant high-performance computing and heuristic requirements. Keywords: Computational biology, Theory of computation, Complexity, High-performance computing, Heuristics Description of the author(s): Davide Cirillo is a postdoctoral researcher at the Computational biology Group within the Life Sciences Department at Barcelona Supercomputing Center (BSC). Davide Cirillo received the MSc degree in Pharmaceutical Biotechnology from University of Rome ‘La Sapienza’, Italy, in 2011, and the PhD degree in biomedicine from Universitat Pompeu Fabra (UPF) and Center for Genomic Regulation (CRG) in Barcelona, Spain, in 2016. His current research interests include Machine Learning, Artificial Intelligence, and Precision medicine. Miguel Ponce de León is a postdoctoral researcher at the Computational biology Group within the Life Sciences Department at Barcelona Supercomputing Center (BSC). Miguel Ponce de León received the MSc degree in bioinformatics from University UdelaR of Uruguay, in 2011, and the PhD degree in Biochemistry and biomedicine from University Complutense de, Madrid (UCM), Spain, in 2017.