'Design of Armadillo Repeat Protein Scaffolds'
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nucleocytoplasmic Shuttling of the Glucocorticoid Receptor Is Influenced by Tetratricopeptide Repeat-Containing Proteins Gisela I
© 2020. Published by The Company of Biologists Ltd | Journal of Cell Science (2020) 133, jcs238873. doi:10.1242/jcs.238873 RESEARCH ARTICLE Nucleocytoplasmic shuttling of the glucocorticoid receptor is influenced by tetratricopeptide repeat-containing proteins Gisela I. Mazaira1,*, Pablo C. Echeverria2,* and Mario D. Galigniana1,3,‡ ABSTRACT FKBP52 (also known as FKBP4), FKBP51 (FKBP5) and CyP40 It has been demonstrated that tetratricopeptide-repeat (TPR) domain (PPID), and the immunophilin-like proteins PP5, FKBPL (WISp39) proteins regulate the subcellular localization of glucocorticoid receptor and FKBP37 (ARA9/XAP2/AIP) are counted among the best (GR). This study analyses the influence of the TPR domain of high characterized members of the TPR domain family of proteins molecular weight immunophilins in the retrograde transport and associated with nuclear receptor family members via Hsp90 (Pratt nuclear retention of GR. Overexpression of the TPR peptide et al., 2004a; Storer et al., 2011). TPR domains are composed of prevented efficient nuclear accumulation of the GR by disrupting the loosely conserved 34 amino acid sequence motifs that are found in formation of complexes with the dynein-associated immunophilin arrays comprising between 2 and 20 repeats, wherein the repeats pack FKBP52 (also known as FKBP4), the adaptor transporter importin-β1 against each other giving rise to coiled and superhelical structures (KPNB1), the nuclear pore-associated glycoprotein Nup62 and nuclear (Perez-Riba and Itzhaki, 2019). Originally identified in components of matrix-associated structures. We also show that nuclear import of GR the anaphase-promoting complex (Sikorski et al., 1991), TPR was impaired, whereas GR nuclear export was enhanced. domains are now known to mediate specific protein interactions in Interestingly, the CRM1 (exportin-1) inhibitor leptomycin-B abolished numerous cellular contexts. -

A Novel LRRK2 Variant P.G2294R in the WD40 Domain Identified in Familial Parkinson's Disease Affects LRRK2 Protein Levels
International Journal of Molecular Sciences Article A Novel LRRK2 Variant p.G2294R in the WD40 Domain Identified in Familial Parkinson’s Disease Affects LRRK2 Protein Levels Jun Ogata 1, Kentaro Hirao 2, Kenya Nishioka 3 , Arisa Hayashida 3, Yuanzhe Li 3, Hiroyo Yoshino 4, Soichiro Shimizu 2, Nobutaka Hattori 1,3,4 and Yuzuru Imai 1,* 1 Department of Research for Parkinson’s Disease, Juntendo University Graduate School of Medicine, Tokyo 113-8421, Japan; [email protected] (J.O.); [email protected] (N.H.) 2 Department of Geriatric Medicine, Tokyo Medical University, 6-7-1 Nishishinjuku, Shinjuku-ku, Tokyo 160-0023, Japan; [email protected] (K.H.); [email protected] (S.S.) 3 Department of Neurology, Juntendo University School of Medicine, 2-1-1 Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; [email protected] (K.N.); [email protected] (A.H.); [email protected] (Y.L.) 4 Research Institute for Diseases of Old Age, Graduate School of Medicine, Juntendo University, 2-1-1 Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-3-6801-8332 Abstract: Leucine-rich repeat kinase 2 (LRRK2) is a major causative gene of late-onset familial Parkin- son’s disease (PD). The suppression of kinase activity is believed to confer neuroprotection, as most pathogenic variants of LRRK2 associated with PD exhibit increased kinase activity. We herein report a novel LRRK2 variant—p.G2294R—located in the WD40 domain, detected through targeted gene- Citation: Ogata, J.; Hirao, K.; panel screening in a patient with familial PD. -
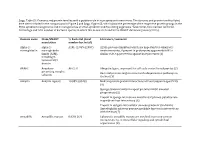
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67) -

Consensus Tetratricopeptide Repeat Proteins Are Complex Superhelical 2 Nanosprings
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.27.437344; this version posted August 30, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Consensus tetratricopeptide repeat proteins are complex superhelical 2 nanosprings 1, ∗ 1 1 2 2 3 Marie Synakewicz, Rohan S. Eapen, Albert Perez-Riba, Daniela Bauer, Andreas Weißl, 3 3 2 1, ∗ 4, ∗ 4 Gerhard Fischer, Marko Hyv¨onen, Matthias Rief, Laura S. Itzhaki, and Johannes Stigler 1 5 Department of Pharmacology, University of Cambridge, 6 Tennis Court Road, Cambridge, CB2 1PD, United Kingdom 2 7 Physik-Department, Technische Universit¨atM¨unchen, 8 James-Franck-Straße 1, 85748 Garching, Germany 3 9 Department of Biochemistry, University of Cambridge, 10 Tennis Court Road, Cambridge, CB2 1GA, United Kingdom 4 11 Gene Center Munich, Ludwig-Maximilians-Universit¨atM¨unchen, 12 Feodor-Lynen-Straße 25, 81377 M¨unchen,Germany 13 (Dated: August 20, 2021) Abstract. Tandem-repeat proteins comprise small secondary structure motifs that stack to form one- dimensional arrays with distinctive mechanical properties that are proposed to direct their cellular functions. Here, we use single-molecule optical tweezers to study the folding of consensus-designed tetratricopeptide repeats (CTPRs) | superhelical arrays of short helix-turn-helix motifs. We find that CTPRs display a spring-like mechanical response in which individual repeats undergo rapid equilibrium fluctuations between folded and unfolded conformations. We rationalise the force response using Ising models and dissect the folding pathway of CTPRs under mechanical load, revealing how the repeat arrays form from the centre towards both termini simultaneously. -

Small Glutamine-Rich Tetratricopeptide Repeat
www.nature.com/scientificreports OPEN Small Glutamine-Rich Tetratricopeptide Repeat- Containing Protein Alpha (SGTA) Received: 04 January 2016 Accepted: 07 June 2016 Ablation Limits Offspring Viability Published: 30 June 2016 and Growth in Mice Lisa K. Philp1, Tanya K. Day1, Miriam S. Butler1, Geraldine Laven-Law1, Shalini Jindal1, Theresa E. Hickey1, Howard I. Scher2, Lisa M. Butler1,3,* & Wayne D. Tilley1,3,* Small glutamine-rich tetratricopeptide repeat-containing protein α (SGTA) has been implicated as a co-chaperone and regulator of androgen and growth hormone receptor (AR, GHR) signalling. We investigated the functional consequences of partial and full Sgta ablation in vivo using Cre-lox Sgta- null mice. Sgta+/− breeders generated viable Sgta−/− offspring, but at less than Mendelian expectancy. Sgta−/− breeders were subfertile with small litters and higher neonatal death (P < 0.02). Body size was significantly and proportionately smaller in male and femaleSgta −/− (vs WT, Sgta+/− P < 0.001) from d19. Serum IGF-1 levels were genotype- and sex-dependent. Food intake, muscle and bone mass and adiposity were unchanged in Sgta−/−. Vital and sex organs had normal relative weight, morphology and histology, although certain androgen-sensitive measures such as penis and preputial size, and testis descent, were greater in Sgta−/−. Expression of AR and its targets remained largely unchanged, although AR localisation was genotype- and tissue-dependent. Generally expression of other TPR- containing proteins was unchanged. In conclusion, this thorough investigation of SGTA-null mutation reports a mild phenotype of reduced body size. The model’s full potential likely will be realised by genetic crosses with other models to interrogate the role of SGTA in the many diseases in which it has been implicated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Eukaryotic Genome Annotation
Comparative Features of Multicellular Eukaryotic Genomes (2017) (First three statistics from www.ensembl.org; other from original papers) C. elegans A. thaliana D. melanogaster M. musculus H. sapiens Species name Nematode Thale Cress Fruit Fly Mouse Human Size (Mb) 103 136 143 3,482 3,555 # Protein-coding genes 20,362 27,655 13,918 22,598 20,338 (25,498 (13,601 original (30,000 (30,000 original est.) original est.) original est.) est.) Transcripts 58,941 55,157 34,749 131,195 200,310 Gene density (#/kb) 1/5 1/4.5 1/8.8 1/83 1/97 LINE/SINE (%) 0.4 0.5 0.7 27.4 33.6 LTR (%) 0.0 4.8 1.5 9.9 8.6 DNA Elements 5.3 5.1 0.7 0.9 3.1 Total repeats 6.5 10.5 3.1 38.6 46.4 Exons % genome size 27 28.8 24.0 per gene 4.0 5.4 4.1 8.4 8.7 average size (bp) 250 506 Introns % genome size 15.6 average size (bp) 168 Arabidopsis Chromosome Structures Sorghum Whole Genome Details Characterizing the Proteome The Protein World • Sequencing has defined o Many, many proteins • How can we use this data to: o Define genes in new genomes o Look for evolutionarily related genes o Follow evolution of genes ▪ Mixing of domains to create new proteins o Uncover important subsets of genes that ▪ That deep phylogenies • Plants vs. animals • Placental vs. non-placental animals • Monocots vs. dicots plants • Common nomenclature needed o Ensure consistency of interpretations InterPro (http://www.ebi.ac.uk/interpro/) Classification of Protein Families • Intergrated documentation resource for protein super families, families, domains and functional sites o Mitchell AL, Attwood TK, Babbitt PC, et al. -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Exploring the Folding Energy Landscape of a Series of Designed Consensus Tetratricopeptide Repeat Proteins
PHYSICS, BIOPHYSICS AND COMPUTATIONAL BIOLOGY BIOPHYSICS AND COMPUTATIONAL BIOLOGY Correction for ‘‘The role of fluctuations and stress on the Correction for ‘‘Exploring the folding energy landscape of a effective viscosity of cell aggregates,’’ by Philippe Marmottant, series of designed consensus tetratricopeptide repeat proteins,’’ Abbas Mgharbel, Jos Ka¨fer,Benjamin Audren, Jean-Paul Rieu, by Yalda Javadi and Ewan R. G. Main, which appeared in issue Jean-Claude Vial, Boudewijn van der Sanden, Athanasius F. M. 41, October 13, 2009, of Proc Natl Acad Sci USA (106:17383– Mare´e, Franc¸ois Graner, and He´le`ne Delanoe¨-Ayari, which 17388; first published October 1, 2009; 10.1073/pnas. appeared in issue 41, October 13, 2009, of Proc Natl Acad Sci 0907455106). USA (106:17271–17275; first published September 25, 2009; The authors note that, due to a printer’s error, some text 10.1073/pnas.0902085106). appeared incorrectly on page 17384. In the left column, the The authors note that, due to a printer’s error, the affiliation second sentence in the third full paragraph, ‘‘This yielded [D]50% for Abbas Mgharbel, Benjamin Audren, Jean-Paul Rieu, and (midpoint of unfolding) and mD-N (change in solvent-accessible ⌬ H2O ⌬ H2O He´le`ne Delanoe¨-Ayari at Universite´ de Lyon appeared incor- surface area upon protein unfolding) from which GD–N/ G03j rectly in part. The name of the authors’ laboratory, Laboratoire (free energy of unfolding in water) was calculated (Table S1),’’ Mate´riaux et Phe´nome`nes Quantiques, should instead have should instead have appeared as ‘‘This yielded [D]50% (midpoint appeared as Laboratoire de Physique de la Matie`re Condense´e of unfolding) and mD-N (change in solvent-accessible surface ⌬ H2O et Nanostructures. -

De Novo PHIP Predicted Deleterious Variants Are Associated with Developmental Delay, Intellectual Disability, Obesity, and Dysmorphic Features
Downloaded from molecularcasestudies.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press De novo PHIP predicted deleterious variants are associated with developmental delay, intellectual disability, obesity, and dysmorphic features Running title: Case study of two patients with PHIP variants Emily Webster1, Megan T. Cho2, Nora Alexander2, Sonal Desai3, Sakkubai Naidu3, Mir Reza Bekheirnia4, Andrea Lewis4, Kyle Retterer2, Jane Juusola2, Wendy K. Chung1,5 1Department of Pediatrics, Columbia University Medical Center, New York, NY, USA 2GeneDx, Gaithersburg, MD, USA 3Kennedy Krieger Institute, Baltimore, MD, USA 4Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA 5Department of Medicine, Columbia University Medical Center, New York, NY, USA Correspondence: Wendy K. Chung, Columbia University Medical Center, 1150 St. Nicholas Avenue, New York, NY 10032, USA, Tel: +1 212 851 5313, Fax: +1 212 851 5306, [email protected] Running title: PHIP variants cause developmental delay and obesity 1 Downloaded from molecularcasestudies.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Using whole exome sequencing, we have identified novel de novo heterozygous Pleckstrin homology domain-interacting protein (PHIP) variants that are predicted to be deleterious, including a frameshift deletion, in two unrelated patients with common clinical features of developmental delay, intellectual disability, anxiety, hypotonia, poor balance, obesity, and dysmorphic features. A nonsense mutation in PHIP has previously been associated with similar clinical features. Patients with microdeletions of 6q14.1 including PHIP have a similar phenotype of developmental delay, intellectual disability, hypotonia, and obesity, suggesting that the phenotype of our patients is a result of loss-of-function mutations. -

Armc5 Deletion Causes Developmental Defects and Compromises T-Cell Immune Responses
ARTICLE Received 26 Jan 2016 | Accepted 4 Nov 2016 | Published 7 Feb 2017 DOI: 10.1038/ncomms13834 OPEN Armc5 deletion causes developmental defects and compromises T-cell immune responses Yan Hu 1,*, Linjiang Lao1,*, Jianning Mao1, Wei Jin1, Hongyu Luo1, Tania Charpentier2, Shijie Qi1, Junzheng Peng1, Bing Hu3, Mieczyslaw Martin Marcinkiewicz4, Alain Lamarre2 & Jiangping Wu1,5 Armadillo repeat containing 5 (ARMC5) is a cytosolic protein with no enzymatic activities. Little is known about its function and mechanisms of action, except that gene mutations are associated with risks of primary macronodular adrenal gland hyperplasia. Here we map Armc5 expression by in situ hybridization, and generate Armc5 knockout mice, which are small in body size. Armc5 knockout mice have compromised T-cell proliferation and differentiation into Th1 and Th17 cells, increased T-cell apoptosis, reduced severity of experimental autoimmune encephalitis, and defective immune responses to lymphocytic choriomeningitis virus infection. These mice also develop adrenal gland hyperplasia in old age. Yeast 2-hybrid assays identify 16 ARMC5-binding partners. Together these data indicate that ARMC5 is crucial in fetal development, T-cell function and adrenal gland growth homeostasis, and that the functions of ARMC5 probably depend on interaction with multiple signalling pathways. 1 Centre de recherche (CR), Centre hospitalier de l’Universite´ de Montre´al (CHUM), 900 Rue Saint Denis, Montre´al, Que´bec, Canada H2X 0A9. 2 Institut national de la recherche scientifique—Institut Armand-Frappier (INRS-IAF), 531 Boul. des Prairies, Laval, Que´bec, Canada H7V 1B7. 3 Anatomic Pathology, AmeriPath Central Florida, 4225 Fowler Ave. Tampa, Orlando, Florida 33617, USA. -

An Unusual Hydrophobic Core Confers Extreme Flexibility to HEAT Repeat Proteins
1596 Biophysical Journal Volume 99 September 2010 1596–1603 An Unusual Hydrophobic Core Confers Extreme Flexibility to HEAT Repeat Proteins Christian Kappel, Ulrich Zachariae, Nicole Do¨lker, and Helmut Grubmu¨ller* Department of Theoretical and Computational Biophysics, Max Planck Institute for Biophysical Chemistry, Go¨ttingen, Germany ABSTRACT Alpha-solenoid proteins are suggested to constitute highly flexible macromolecules, whose structural variability and large surface area is instrumental in many important protein-protein binding processes. By equilibrium and nonequilibrium molecular dynamics simulations, we show that importin-b, an archetypical a-solenoid, displays unprecedentedly large and fully reversible elasticity. Our stretching molecular dynamics simulations reveal full elasticity over up to twofold end-to-end extensions compared to its bound state. Despite the absence of any long-range intramolecular contacts, the protein can return to its equilibrium structure to within 3 A˚ backbone RMSD after the release of mechanical stress. We find that this extreme degree of flexibility is based on an unusually flexible hydrophobic core that differs substantially from that of structurally similar but more rigid globular proteins. In that respect, the core of importin-b resembles molten globules. The elastic behavior is dominated by nonpolar interactions between HEAT repeats, combined with conformational entropic effects. Our results suggest that a-sole- noid structures such as importin-b may bridge the molecular gap between completely structured and intrinsically disordered proteins. INTRODUCTION Solenoid proteins, consisting of repeating arrays of simple on repeat proteins focus on the folding and unfolding basic structural motifs, account for >5% of the genome of mechanism (14–16), an atomic force microscopy study on multicellular organisms (1).