Downexpression of HSD17B6 Correlates with Clinical Prognosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Analysis of Retinopathy in Type 1 Diabetes
Genetic Analysis of Retinopathy in Type 1 Diabetes by Sayed Mohsen Hosseini A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Institute of Medical Science University of Toronto © Copyright by S. Mohsen Hosseini 2014 Genetic Analysis of Retinopathy in Type 1 Diabetes Sayed Mohsen Hosseini Doctor of Philosophy Institute of Medical Science University of Toronto 2014 Abstract Diabetic retinopathy (DR) is a leading cause of blindness worldwide. Several lines of evidence suggest a genetic contribution to the risk of DR; however, no genetic variant has shown convincing association with DR in genome-wide association studies (GWAS). To identify common polymorphisms associated with DR, meta-GWAS were performed in three type 1 diabetes cohorts of White subjects: Diabetes Complications and Control Trial (DCCT, n=1304), Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR, n=603) and Renin-Angiotensin System Study (RASS, n=239). Severe (SDR) and mild (MDR) retinopathy outcomes were defined based on repeated fundus photographs in each study graded for retinopathy severity on the Early Treatment Diabetic Retinopathy Study (ETDRS) scale. Multivariable models accounted for glycemia (measured by A1C), diabetes duration and other relevant covariates in the association analyses of additive genotypes with SDR and MDR. Fixed-effects meta- analysis was used to combine the results of GWAS performed separately in WESDR, ii RASS and subgroups of DCCT, defined by cohort and treatment group. Top association signals were prioritized for replication, based on previous supporting knowledge from the literature, followed by replication in three independent white T1D studies: Genesis-GeneDiab (n=502), Steno (n=936) and FinnDiane (n=2194). -

Exploring the Pharmacological Mechanism of Quercetin-Resveratrol Combination for Polycystic Ovary Syndrome
www.nature.com/scientificreports Corrected: Publisher Correction OPEN Exploring the Pharmacological Mechanism of Quercetin- Resveratrol Combination for Polycystic Ovary Syndrome: A Systematic Pharmacological Strategy-Based Research Kailin Yang1,2,6, Liuting Zeng 3,6*, Tingting Bao3,4,6, Zhiyong Long5 & Bing Jin3* Resveratrol and quercetin have efects on polycystic ovary syndrome (PCOS). Hence, resveratrol combined with quercetin may have better efects on it. However, because of the limitations in animal and human experiments, the pharmacological and molecular mechanism of quercetin-resveratrol combination (QRC) remains to be clarifed. In this research, a systematic pharmacological approach comprising multiple compound target collection, multiple potential target prediction, and network analysis was used for comparing the characteristic of resveratrol, quercetin and QRC, and exploring the mechanism of QRC. After that, four networks were constructed and analyzed: (1) compound-compound target network; (2) compound-potential target network; (3) QRC-PCOS PPI network; (4) QRC-PCOS- other human proteins (protein-protein interaction) PPI network. Through GO and pathway enrichment analysis, it can be found that three compounds focus on diferent biological processes and pathways; and it seems that QRC combines the characteristics of resveratrol and quercetin. The in-depth study of QRC further showed more PCOS-related biological processes and pathways. Hence, this research not only ofers clues to the researcher who is interested in comparing the diferences among resveratrol, quercetin and QRC, but also provides hints for the researcher who wants to explore QRC’s various synergies and its pharmacological and molecular mechanism. Polycystic ovary syndrome (PCOS) is one of the most common female endocrine diseases characterized by hyperandrogenism, menstrual disorders and infertility. -

Transdifferentiation of Human Mesenchymal Stem Cells
Transdifferentiation of Human Mesenchymal Stem Cells Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Julius-Maximilians-Universität Würzburg vorgelegt von Tatjana Schilling aus San Miguel de Tucuman, Argentinien Würzburg, 2007 Eingereicht am: Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Martin J. Müller Gutachter: PD Dr. Norbert Schütze Gutachter: Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am: Hiermit erkläre ich ehrenwörtlich, dass ich die vorliegende Dissertation selbstständig angefertigt und keine anderen als die von mir angegebenen Hilfsmittel und Quellen verwendet habe. Des Weiteren erkläre ich, dass diese Arbeit weder in gleicher noch in ähnlicher Form in einem Prüfungsverfahren vorgelegen hat und ich noch keinen Promotionsversuch unternommen habe. Gerbrunn, 4. Mai 2007 Tatjana Schilling Table of contents i Table of contents 1 Summary ........................................................................................................................ 1 1.1 Summary.................................................................................................................... 1 1.2 Zusammenfassung..................................................................................................... 2 2 Introduction.................................................................................................................... 4 2.1 Osteoporosis and the fatty degeneration of the bone marrow..................................... 4 2.2 Adipose and bone -

Identification of Genes Associated with Castration‑Resistant Prostate Cancer by Gene Expression Profile Analysis
MOLECULAR MEDICINE REPORTS 16: 6803-6813, 2017 Identification of genes associated with castration‑resistant prostate cancer by gene expression profile analysis CHUI GUO HUANG1, FENG XI LI2, SONG PAN3, CHANG BAO XU1, JUN QIANG DAI4 and XING HUA ZHAO1 1Department of Urology, The Second Affiliated Hospital, Zhengzhou University, Zhengzhou, Henan 450014; 2Department of Gastrointestinal Glands Surgery, The First Affiliated Hospital, Guangxi Medical University, Nanning, Guangxi 530000; 3Department of Urology, The First Affiliated Hospital, Zhengzhou University, Zhengzhou, Henan 450014; 4Department of Neurosurgery, The Second Affiliated Hospital, Lanzhou University, Lanzhou, Gansu 730030, P.R. China Received December 16, 2016; Accepted July 17, 2017 DOI: 10.3892/mmr.2017.7488 Abstract. Prostate cancer (CaP) is a serious and common western countries (1). CaP growth is driven by androgens; genital tumor. Generally, men with metastatic CaP can easily therefore, the conventional treatment option is to lower the develop castration-resistant prostate cancer (CRPC). However, levels of male sex hormones (2). Current treatment strate- the pathogenesis and tumorigenic pathways of CRPC remain to gies for CaP include surgery, external beam radiotherapy, be elucidated. The present study performed a comprehensive brachytherapy, chemotherapy and androgen-deprivation analysis on the gene expression profile of CRPC in order to therapy (ADT) (2,3). ADT often involves surgically determine the pathogenesis and tumorigenic of CRPC. The removing the testicles or using drugs to block the andro- GSE33316 microarray, which consisted of 5 non-castrated gens from affecting the body (4). Unfortunately, ADT has samples and 5 castrated samples, was downloaded from the gene a limited role in preventing majority of patients progressing expression omnibus database. -

Lethal Congenital Contractural Syndrome Type 2 (LCCS2) Is Caused by a Mutation in ERBB3 (Her3), a Modulator of the Phosphatidylinositol-3-Kinase/Akt Pathway
REPORT Lethal Congenital Contractural Syndrome Type 2 (LCCS2) Is Caused by a Mutation in ERBB3 (Her3), a Modulator of the Phosphatidylinositol-3-Kinase/Akt Pathway Ginat Narkis, Rivka Ofir, Esther Manor, Daniella Landau, Khalil Elbedour, and Ohad S. Birk Lethal congenital contractural syndrome type 2 (LCCS2) is an autosomal recessive neurogenic form of arthrogryposis that is associated with atrophy of the anterior horn of the spinal cord. We previously mapped LCCS2 to 6.4 Mb on chromosome 12q13 and have now narrowed the locus to 4.6 Mb. We show that the disease is caused by aberrant splicing of ERBB3, which leads to a predicted truncated protein. ERBB3 (Her3), an activator of the phosphatidylinositol-3-kinase/ Akt pathway—regulating cell survival and vesicle trafficking—is essential for the generation of precursors of Schwann cells that normally accompany peripheral axons of motor neurons. Gain-of-function mutations in members of the epidermal growth-factor tyrosine kinase–receptor family have been associated with predilection to cancer. This is the first report of a human phenotype resulting from loss of function of a member of this group. Lethal congenital contractual syndrome type 2 (LCCS2 shown). The possibility that the disease-associated locus [MIM 607598]), a neonatally lethal form of arthrogryposis is between D12S305 and D12S1072 cannot be ruled out, presenting in two Israeli-Bedouin kindreds1 (families A but it is less likely, since this would require two crossing- and B) (fig. 1), is characterized by multiple joint contrac- over events occurring in individual 61 (fig. 2). tures, anterior horn atrophy in the spinal cord, and a In an attempt to identify the specific molecular defect unique feature of a markedly distended urinary bladder. -

Anwendung Der Mikroarray-Technologie Für Eine Globale Charakterisierung Der Spermatogenese Des Menschen Und Seiner Pathologien
Anwendung der Mikroarray-Technologie für eine globale Charakterisierung der Spermatogenese des Menschen und seiner Pathologien Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften im Fachbereich Chemie der Universität Hamburg vorgelegt von CAROLINE FEIG Hamburg 2007 Die vorliegende Arbeit wurde in der Abteilung für Andrologie des Universitätsklinikums Hamburg-Eppendorf durchgeführt. 1. Dissertationsgutachter: Herr Prof. Dr. Ulrich Hahn Abteilung für Biochemie und Molekularbiologie Martin-Luther-King-Platz 6 20146 Hamburg 2. Dissertationsgutachterin: Frau Prof. Dr. Christiane Kirchhoff Abteilung für Andrologie Universitätsklinikum Hamburg-Eppendorf Martinistrasse 52 20246 Hamburg Tag der Disputation: 11. Mai 2007 Inhaltsverzeichnis I Inhaltsverzeichnis ................................................................................................ I Abkürzungsverzeichnis ..................................................................................... V 1 Einleitung ............................................................................................................ 1 1.1 Das Problem der männlichen Infertilität................................................................. 1 1.2 Die Organisation des Hodens.................................................................................. 3 1.3 Die Spermatogenese................................................................................................ 4 1.3.1 Phasen der Spermatogenese ................................................................................... -

Mitotic Checkpoints and Chromosome Instability Are Strong Predictors of Clinical Outcome in Gastrointestinal Stromal Tumors
MITOTIC CHECKPOINTS AND CHROMOSOME INSTABILITY ARE STRONG PREDICTORS OF CLINICAL OUTCOME IN GASTROINTESTINAL STROMAL TUMORS. Pauline Lagarde1,2, Gaëlle Pérot1, Audrey Kauffmann3, Céline Brulard1, Valérie Dapremont2, Isabelle Hostein2, Agnès Neuville1,2, Agnieszka Wozniak4, Raf Sciot5, Patrick Schöffski4, Alain Aurias1,6, Jean-Michel Coindre1,2,7 Maria Debiec-Rychter8, Frédéric Chibon1,2. Supplemental data NM cases deletion frequency. frequency. deletion NM cases Mand between difference the highest setswith of theprobe a view isdetailed panel Bottom frequently. sorted totheless deleted theprobe are frequently from more and thefrequency deletion represent Yaxes inblue. are cases (NM) metastatic for non- frequencies Corresponding inmetastatic (red). probe (M)cases sets figureSupplementary 1: 100 100 20 40 60 80 20 40 60 80 0 0 chr14 1 chr14 88 chr14 175 chr14 262 chr9 -MTAP 349 chr9 -MTAP 436 523 chr9-CDKN2A 610 Histogram presenting the 2000 more frequently deleted deleted frequently the 2000 more presenting Histogram chr9-CDKN2A 697 chr9-CDKN2A 784 chr9-CDKN2B 871 chr9-CDKN2B 958 chr9-CDKN2B 1045 chr22 1132 chr22 1219 chr22 1306 chr22 1393 1480 1567 M NM 1654 1741 1828 1915 M NM GIST14 GIST2 GIST16 GIST3 GIST19 GIST63 GIST9 GIST38 GIST61 GIST39 GIST56 GIST37 GIST47 GIST58 GIST28 GIST5 GIST17 GIST57 GIST47 GIST58 GIST28 GIST5 GIST17 GIST57 CDKN2A Supplementary figure 2: Chromosome 9 genomic profiles of the 18 metastatic GISTs (upper panel). Deletions and gains are indicated in green and red, respectively; and color intensity is proportional to copy number changes. A detailed view is given (bottom panel) for the 6 cases presenting a homozygous 9p21 deletion targeting CDKN2A locus (dark green). -

Title: EHCO: Encyclopedia of Hepatocellular Carcinoma Genes Online
Title: Detection of the inferred interaction network in hepatocellular carcinoma from EHCO (Encyclopedia of Hepatocellular Carcinoma genes Online) Chun-Nan Hsu1*, Jin-Mei Lai2*, Chia-Hung Liu1*, Huei-Hun Tseng3*, Chih-Yun Lin4*, Kuan-Ting Lin1*, Hsu-Hua Yeh3, Ting-Yi Sung1, Wen-Lian Hsu1, Li-Jen Su4, Sheng-An Lee4, 5, Chang-Han Chen3, 4, Gen-Cher Lee1, 5, D. T. Lee1, 5, Yow-Ling Shiue6, Chang-Wei Yeh7, Chao-Hui Chang4, Cheng-Yan Kao5, Chi-Ying F. Huang3, 4, 5, 8,9§ 1Institute of Information Science, Academia Sinica, Taipei 115, Taiwan, R. O. C. 2Department of Life Science, Fu-Jen Catholic University, Taipei Hsien 242, Taiwan, R. O. C. 3Division of Molecular and Genomic Medicine, National Health Research Institutes, Miaoli County 350, Taiwan, R. O. C. 4Institute of Cancer Research, National Health Research Institutes, Taipei 114, Taiwan, R. O. C. 5Department of Computer Science and Information Engineering, National Taiwan University, Taipei 106, Taiwan, R. O. C. 6Institute of Biomedical Science, National Sun Yat-Sen University, Kaohsiung 804, Taiwan, R. O. C. 7National Center for High-performance Computing, Hsinchu 300, Taiwan, R. O. C. 8Institute of Bio-Pharmaceutical Sciences and 9Institute of Biotechnology in Medicine, National Yang-Ming University, Taipei 112, Taiwan, R. O. C. *These authors contribute equally to this work. §Corresponding author Chi-Ying F. Huang, Ph.D. Associate Investigator Institute of Cancer Research National Health Research Institutes No. 161, Sec. 6, Min-Chuan East Road Taipei 114, Taiwan, R.O.C. Phone: (886)-2-26534401 -

Two Pathways of Fetal Testicular Androgen Biosynthesis Are Needed for Male Sexual Differentiation
ARTICLE Why Boys Will Be Boys: Two Pathways of Fetal Testicular Androgen Biosynthesis Are Needed for Male Sexual Differentiation Christa E. Flu¨ck,1,* Monika Meyer-Bo¨ni,2 Amit V. Pandey,1 Petra Kempna´,1 Walter L. Miller,3 Eugen J. Schoenle,2 and Anna Biason-Lauber2,* Human sexual determination is initiated by a cascade of genes that lead to the development of the fetal gonad. Whereas development of the female external genitalia does not require fetal ovarian hormones, male genital development requires the action of testicular testos- terone and its more potent derivative dihydrotestosterone (DHT). The ‘‘classic’’ biosynthetic pathway from cholesterol to testosterone in the testis and the subsequent conversion of testosterone to DHT in genital skin is well established. Recently, an alternative pathway leading to DHT has been described in marsupials, but its potential importance to human development is unclear. AKR1C2 is an enzyme that participates in the alternative but not the classic pathway. Using a candidate gene approach, we identified AKR1C2 mutations with sex-limited recessive inheritance in four 46,XY individuals with disordered sexual development (DSD). Analysis of the inheritance of microsatellite markers excluded other candidate loci. Affected individuals had moderate to severe undervirilization at birth; when recre- ated by site-directed mutagenesis and expressed in bacteria, the mutant AKR1C2 had diminished but not absent catalytic activities. The 46,XY DSD individuals also carry a mutation causing aberrant splicing in AKR1C4, which encodes an enzyme with similar activity. This suggests a mode of inheritance where the severity of the developmental defect depends on the number of mutations in the two genes. -
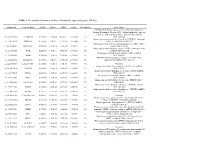
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137. -

Key Genes Involved in Cell Cycle Arrest and DNA Damage Repair Identified in Anaplastic Thyroid Carcinoma Using Integrated Bioinformatics Analysis
4203 Original Article Key genes involved in cell cycle arrest and DNA damage repair identified in anaplastic thyroid carcinoma using integrated bioinformatics analysis Zhi Zhang1#, Zhenning Zou2#, Haixia Dai3#, Ruifang Ye2#, Xiaoqing Di4, Rujia Li2, Yanping Ha2, Yanqin Sun2, Siyuan Gan2 1Department of Thyroid and Mammary Vascular Surgery, the Affiliated Hospital of Guangdong Medical University, Zhanjiang, China; 2Department of Pathology, Guangdong Medical University, Zhanjiang, China; 3Department of Ultrasound, 4Department of Pathology, the Affiliated Hospital of Guangdong Medical University, Zhanjiang, China Contributions: (I) Conception and design: S Gan, Z Zhang, Y Sun; (II) Administrative support: S Gan, Y Sun; (III) Provision of study materials or patients: S Gan, Z Zou, H Dai, R Ye; (IV) Collection and assembly of data: S Gan, X Di, R Li, Y Ha; (V) Data analysis and interpretation: S Gan, Z Zhang, Y Sun, Z Zou, H Dai; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. #These authors contributed equally to this work. Correspondence to: Siyuan Gan; Yanqin Sun. Department of Pathology, Guangdong Medical University, Xiashan District, Zhanjiang, China. Email: [email protected]; [email protected]. Background: Since anaplastic thyroid carcinoma (ATC) has rapid progression and a poor outcome, identification of the key genes and underlying mechanisms of ATC is required. Methods: Gene expression profiles of GSE29265 and GSE33630 were available from the Gene Expression Omnibus database. The two profile datasets included 19 ATC tissues, 55 normal thyroid tissues and 59 papillary thyroid cancer (PTC) tissues. Differentially expressed genes (DEGs) between ATC tissues and normal thyroid tissues as well as ATC tissues and PTC tissues were identified using the GEO2R tool. -

Neuron, Volume 62 Supplemental Data Functional and Evolutionary
Neuron, Volume 62 Supplemental Data Functional and Evolutionary Insights into Human Brain Development Through Global Transcriptome Analysis Matthew B. Johnson, Yuka Imamura Kawasawa, Christopher E. Mason, Željka Krsnik, Giovanni Coppola, Darko Bogdanović, Daniel H. Geschwind, Shrikant M. Mane, Matthew W. State, and Nenad Šestan Supplemental Material: Supplemental Figures Supplemental Experimental Procedures Supplemental References Supplemental Tables Supplemental Figures Supplemental Figure 1. Normal Cytoarchitecture of Brain Specimens Used for Exon Array Analysis Nissl staining of the tissue remaining after microdissection from all four brains used for microarray analysis (specimens 1-4), confirming normal cytoarchitecture and the absence of microscopic neuropathological defects such as periventricular lesions commonly present at this developmental stage. MZ, marginal zone; CP, cortical plate; SP, subplate; SVZ, subventricular zone; VZ, ventricular zone. Scale bar, 250 µm. 2 Supplemental Figure 2. Normal Laminar Position of Cortical Neurons in Brain Specimens Used for Exon Array Analysis Various immunohistochemical markers were used to confirm the presence of all major neuronal and glial cell types present at this developmental age in all four brains. Shown here are selected examples of immunohistochemical staining for markers of layer selective of projection neurons (FOXP2, SOX5, BCL11B, and POU3F3), MZ Cajal-Retzius neurons (RELN) and interneurons (GABA) in the neocortex of the 19 wg brain. The normal laminar position of these neurons indicates absence of obvious defects in neuronal specification and migration. Scale bar, 250 µm. 3 Supplemental Figure 3. Predicted Rates of Copy Number Variation in Brain Specimens Used for Exon Array Analysis Illumina genotyping microarray data were analyzed for putative copy number variations (CNVs) using the PennCNV algorithm (Wang et al., 2007).