(Genus Myuchelys), with Special Reference to the Endangered M. Bellii
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Geographic Variation in the Matamata Turtle, Chelus Fimbriatus, with Observations on Its Shell Morphology and Morphometry
n*entilkilt ilil Biok,gr', 1995. l(-l):19: 1995 by CheloninD Research Foundltion Geographic Variation in the Matamata Turtle, Chelus fimbriatus, with Observations on its Shell Morphology and Morphometry MlncBLo R. SANcnnz-Vu,urcnAr, PnrER C.H. PnrrcHARD:, ArrnEro P.rorrLLo-r, aNn Onan J. LINlnBs3 tDepartment of Biological Anthropolog-,- and Anatomy, Duke lJniversin' Medical Cetter. Box 3170, Dtu'hcun, North Carolina277l0 USA IFat 919-684-8034]; 2Florida Audubotr Societ-t, 460 High,n;a,- 436, Suite 200, Casselberry, Florida 32707 USA: iDepartanento de Esttdios Anbientales, llniyersitlad Sinzrin Bolltnt", Caracas ]O80-A, APDO 89OOO l/enerte\a Ansrucr. - A sample of 126 specimens of Chelusftmbriatus was examined for geographic variation and morphology of the shell. A high degree of variation was found in the plastral formula and in the shape and size of the intergular scute. This study suggests that the Amazon population of matamatas is different from the Orinoco population in the following characters: shape ofthe carapace, plastral pigmentation, and coloration on the underside of the neck. Additionatly, a preliminary analysis indicates that the two populations could be separated on the basis of the allometric growth of the carapace in relation to the plastron. Kry Wonus. - Reptilia; Testudinesl Chelidae; Chelus fimbriatus; turtle; geographic variationl allometryl sexual dimorphism; morphology; morphometryl osteology; South America 'Ihe matamata turtle (Chelus fimbricttus) inhabits the scute morpholo..ey. Measured characters (in all cases straight- Amazon, Oyapoque. Essequibo. and Orinoco river systems line) were: maximum carapace len.-uth (CL). cArapace width of northern South America (Iverson. 1986). Despite a mod- at the ler,'el of the sixth marginal scute (CW). -

Influence of Habitat and Predation on Population Dynamics of the Freshwater Turtle Myuchelys Georgesi
Herpetologica, 69(1), 2013, 46–57 Ó 2013 by The Herpetologists’ League, Inc. INFLUENCE OF HABITAT AND PREDATION ON POPULATION DYNAMICS OF THE FRESHWATER TURTLE MYUCHELYS GEORGESI 1,2,4 1,3 SEAN J. BLAMIRES AND RICKY-JOHN SPENCER 1School of Biological Sciences A08, University of Sydney, NSW 2006, Australia. 2Department of Life Science, Tunghai University, 181 Section 3, Taichung-kan Road, Taichung City, Taiwan 407-04, R.O.C. 3School of Science and Health, Wildlife and Aquatic Ecology Group (Native and Pest Animal Unit), University of Western Sydney, Locked Bag 1797, Penrith NSW 2751, Australia ABSTRACT: Demographic models identify whether animals are vulnerable to local extirpation, but including all ecological parameters across life history stages may be impeded by practical difficulties. When processes acting on certain life stages cannot be measured, extrapolations are often made. A previous study documented that the range of the turtle Myuchelys georgesi is restricted to the Bellinger River, New South Wales, Australia, and its population is stable. We assessed whether M. georgesi selects certain habitats by comparing their distribution among different water holes. We assessed the threat of catfish predation by examining the stomach contents of catfish specimens. We then evaluated whether threats to M. georgesi were likely to have been underestimated by extending our previous demographic model. We did this by revising the previous estimates of adult, juvenile, and hatchling survivorship under hypothetical variations in water hole use and in the presence or absence of catfish predators. We found that M. georgesi preferentially uses moderate to deep water holes. We also found that although catfish 250À400 mm consume hatchling or juvenile turtles, those . -

Download Vol. 33, No. 3
1. , F -6 ~.: 1/JJ/im--3'PT* JL* iLLLZW 1- : s . &, , I ' 4% Or *-* 0 4 Z 0 8 of the FLORIDA STATE MUSEUM Biological Sciences Volume 33 1988 Number 3 REPRODUCTIVE STRATEGIES OF SYMPATRIC FRESHWATER EMYDID TURTLES IN NORTHERN PENINSULAR FLORIDA Dale R. Jackson 3-C p . i ... h¢ 4 f .6/ I Se 4 .¢,$ I - - 64». 4 +Ay. 9.H>« UNIVERSITY OF FLORIDA GAINESVILLE Numbers of the BULLETIN OF TIIE FLORIDA STATE MUSEUM, BIOLOGICAL SCIENCES, are published at irregular intervals. Volumes contain about 300 pages and,are not necessarily completed in any one calendar year. S. DAVID WEBB, Editor OLIVER L. AUSTIN, JR., Editor Bile,ints RHODA J. BRYANT, Managing Editor Communications concerning purchase or exchange of the publications and all manuscripts should be addressed to: Managing Editor, Bulletin; Florida State Museum; University of Florida; Gainesville FL 32611; U.S.A. This public document was promulgated at an annual cost of $2003.53 or $2.000 per copy. It makes available to libraries, scholars, and all interested persons the results of researches in the natural sciences, emphasizing the circum- Caribbean region. ISSN: 0071-6154 CODEN: BFSBAS Publication date: 8/27 Price: $2.00 REPRODUCTIVE STRATEGIES OF SYMPATRIC FRESHWATER EMYDID TURTLES IN NORTHERN PENINSULAR FLORIDA Dale R. Jackson Frontispiece. Alligator nest on Payne's Prairie, Alachua County, Florida, opened to expose seven clutches of Psmdenzys nelsoni eggs and one clutch of Trioi,br ferox eggs (far lower right) surrounding the central clutch of alligator eggs. Most of the alligator eggs had been destroyed earlier by raccoons. REPRODUCTIVE STRATEGIES OF SYMPATRIC FRESHWATER EMYDID TURTLES IN NORTHERN PENINSULAR FLORIDA Dale R. -

Haemogregariny Parazitující U Želv Rodu Pelusios: Fylogenetické Vztahy, Morfologie a Hostitelská Specifita
Haemogregariny parazitující u želv rodu Pelusios: fylogenetické vztahy, morfologie a hostitelská specifita Diplomová práce Bc. Aneta Maršíková Školitel: MVDr. Jana Kvičerová, Ph.D. Školitel specialista: doc. MVDr. Pavel Široký, Ph.D. České Budějovice 2016 Maršíková A., 2016: Haemogregariny parazitující u želv rodu Pelusios: fylogenetické vztahy, morfologie a hostitelská specifita. [Haemogregarines in Pelusios turtles: phylogenetic relationships, morphology, and host specificity, MSc. Thesis, in Czech] – 68 pp., Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic. Annotation: The study deals with phylogenetic relationships, morphology and host specificity of blood parasites Haemogregarina sp. infecting freswater turtles of the genus Pelusios from Africa. Results of phylogenetic analyses are also used for clarification of phylogenetic relationships between "haemogregarines sensu lato" and genus Haemogregarina. Prohlašuji, že svoji diplomovou práci jsem vypracovala samostatně pouze s použitím pramenů a literatury uvedených v seznamu citované literatury. Prohlašuji, že v souladu s § 47b zákona č. 111/1998 Sb. v platném znění souhlasím se zveřejněním své bakalářské práce, a to v nezkrácené podobě – v úpravě vzniklé vypuštěním vyznačených částí archivovaných Přírodovědeckou fakultou - elektronickou cestou ve veřejně přístupné části databáze STAG provozované Jihočeskou univerzitou v Českých Budějovicích na jejích internetových stránkách, a to se zachováním mého autorského práva k odevzdanému textu této kvalifikační práce. Souhlasím dále s tím, aby toutéž elektronickou cestou byly v souladu s uvedeným ustanovením zákona č. 111/1998 Sb. zveřejněny posudky školitele a oponentů práce i záznam o průběhu a výsledku obhajoby kvalifikační práce. Rovněž souhlasím s porovnáním textu mé kvalifikační práce s databází kvalifikačních prací Theses.cz provozovanou Národním registrem vysokoškolských kvalifikačních prací a systémem na odhalování plagiátů. -
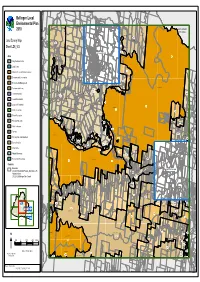
LEP 2010 LZN Template
WILD CATTLE CREEK E1E1E1 DORRIGO RU2RU2 GLENREAGH MULDIVA RU2RU2 OLD BILLINGS RD RAILWAY NATURE COAST TYRINGHAM RESERVE RD RD Bellingen Local E1EE1E11 BORRA CREEK DORRIGO NATIONAL PARK CORAMBA RD E3EE3E33 SLINGSBYS Environmental Plan LITTLE MURRAY RIVER RU2RRU2U2 RD LITTLE PLAIN CREEK BORRA CREEK TYRINGHAM COFFSCOFFS HARBOURHARBOUR 2010 RD CITYCCITYITY COUNCILCOUNCIL BREAKWELLS WILD CATTLE CREEK RD RU2RRU2U2 CORAMBA RD RU2RU2 NEAVES NEAVES RD RD SLINGSBYS RD Land Zoning Map LITTLE MURRAY RIVER OLD COAST RD E3EE3E33 RU2RRU2U2 Sheet LZN_004 E3EE3E33 DEER VALE RD OLD CORAMBA TYRINGHAM RD NTH RD ROCKY CREEK RU1RU1 BARTLETTS RD ReferRefer toto mapmap LZN_004ALZN_004A Zone RU1RRU1U1 BIELSDOWN RIVER E1EE1E11 DEER VALE RD B1 Neighbourhood Centre RU1RU1 DORRIGO NATIONAL PARK B2 Local Centre JOHNSENS WATERFALL WAY OLD CORAMBA WATERFALL WAY RD STH EE3E3E33 E1 National Parks and Nature Reserves E3E3E3 BENNETTS RD E2 Environmental Conservation WATERFALL WAY SHEPHERDS RD RU1RRU1U1 E3 Environmental Management DOME RD EVERINGHAMS ROCKY CREEK RD BIELSDOWN RIVER DORRIGO NATIONAL PARK E4 Environmental Living E3EE3E33 SP1SP1SP1 E3E3E3 SHEPHERDS RD WHISKY CEMETERYCCEMETERYEMETERY DOME RD IN1 General Industrial CREEK RD RU1RRU1U1 RU1RRU1U1 WOODLANDS RD WATERFALL R1 General Residential WAY RU1RRU1U1 PROMISED RU1RU1 E3E3E3 LAND RD WATERFALL WAY R5 Large Lot Residential WHISKY CREEK E1EE1E11 RU1RU1 DORRIGO NATIONAL PARK E1E1E1 E1EE1E11 RE1 Public Recreation BIELSDOWN RIVER E3EE3E33 ROCKY CREEK RD RE2 Private Recreation WHISKY CREEK RD RU1RRU1U1 RU2RU2 E3E3E3 -

Natural Values and Resource Use in the Limmen Bight
NATURAL VALUES AND RESOURCE USE IN THE LIMMEN BIGHT REGION © Australian Marine Conservation Society, January 2019 Australian Marine Conservation Society Phone: +61 (07) 3846 6777 Freecall: 1800 066 299 Email: [email protected] PO Box 5815 West End QLD 4101 Keep Top End Coasts Healthy Alliance Keep Top End Coasts Healthy is an alliance of environment groups including the Australian Marine Conservation Society, the Pew Charitable Trusts and the Environment Centre of the Northern Territory. Authors: Chris Smyth and Joel Turner, Centre for Conservation Geography Printing: Printed on 100% recycled paper by IMAGE OFFSET, Darwin. Maps: Centre for Conservation Geography This report is an independent research paper prepared by the Centre for Conservation Geography commissioned by, and for the exclusive use of, the Keep Top End Coasts Healthy (KTECH) alliance. The report must only be used by KTECH, or with the explicit permission of KTECH. The matters covered in the report are those agreed to between KTECH and the authors. The report does not purport to consider exhaustively all values of the Limmen Bight region. The authors do not accept liability for any loss or damage, including without limitation, compensatory, direct, indirect, or consequential damages and claims of third parties that may be caused directly or indirectly through the use of, reliance upon or interpretation of the contents of the report. Cover photos: Main - Limmen River. Photo: David Hancock Inset (L-R): Green Turtle, Recreational fishing is an important leisure activity in -

Springs of the Mataranka Area 4
THE BIG PICTURE SPRINGS OF THE MATARANKA AREA 4. THE SWAMP Timor Sea A cavernous limestone aquifer extends The large swampy area located on the south side of across a large part of the Northern Territory the Roper River also owes it’s existence to a and into Queensland. The springs at The Mataranka area is notable for its many springs. geological structure that has caused the aquifer to DARWIN Mataranka are one of several outlet points for become shallower and to thin out towards the Roper the aquifer. Other big springs are found on 295000mE This map shows the location of the main springs and other groundwater discharge features. River (see the cross-section). This has resulted in a the Flora, Katherine and Daly Rivers and in broad area underlain by a shallow watertable and Queensland on the Lawn Hill Creek and It explains why the springs are there and describes some of their characteristics. zones of seepage. Extensive tufa deposits formed there because a ridge of bedrock located downstream Daly River Springs KATHERINE Gulf Gregory River. At Mataranka the water of originates from areas to the southeast as far of the seepage zone, ponded the water in a similar Flora River Springs MATARANKA Carpentaria away as the Barkly Tablelands and from the manner to the rock bars in streams as described in 300000mE northwest as far as the King River. Qa1 the note on tufa formation. The ridge formed a base for tufa to accumulate. Tufa dams merged and grew - Waterhouse Cmt over older ones, eventually forming a continuous sheet of limestone. -

Demographic Consequences of Superabundance in Krefft's River
i The comparative ecology of Krefft’s River Turtle Emydura krefftii in Tropical North Queensland. By Dane F. Trembath B.Sc. (Zoology) Applied Ecology Research Group University of Canberra ACT, 2601 Australia A thesis submitted in fulfilment of the requirements of the degree of Masters of Applied Science (Resource Management). August 2005. ii Abstract An ecological study was undertaken on four populations of Krefft’s River Turtle Emydura krefftii inhabiting the Townsville Area of Tropical North Queensland. Two sites were located in the Ross River, which runs through the urban areas of Townsville, and two sites were in rural areas at Alligator Creek and Stuart Creek (known as the Townsville Creeks). Earlier studies of the populations in Ross River had determined that the turtles existed at an exceptionally high density, that is, they were superabundant, and so the Townsville Creek sites were chosen as low abundance sites for comparison. The first aim of this study was to determine if there had been any demographic consequences caused by the abundance of turtle populations of the Ross River. Secondly, the project aimed to determine if the impoundments in the Ross River had affected the freshwater turtle fauna. Specifically this study aimed to determine if there were any difference between the growth, size at maturity, sexual dimorphism, size distribution, and diet of Emydura krefftii inhabiting two very different populations. A mark-recapture program estimated the turtle population sizes at between 490 and 5350 turtles per hectare. Most populations exhibited a predominant female sex-bias over the sampling period. Growth rates were rapid in juveniles but slowed once sexual maturity was attained; in males, growth basically stopped at maturity, but in females, growth continued post-maturity, although at a slower rate. -

Bellinger and Kalang River Estuaries Erosion Study
Bellinger and Kalang River Estuaries Erosion Study Damon Telfer GECO Environmental Tim Cohen IRM Consultants February 2010 Report prep ared for BELLINGEN SHIRE COUNCIL DISCLAIMER Whilst every care has been taken in compiling this erosion study and report, no assurances are given that it is free from error or omission. The authors make no expressed or implied warranty of the accuracy or fitness of the recommendations in the report and disclaim all liability for the consequences of anything done or omitted to be done by any person in reliance upon these recommendations. The use of this report is at the user’s risk and discretion. The authors will not be liable for any loss, damage, costs or injury including consequential, incidental, or financial loss arising out of the use of this report. This report was produced with financial assistance from the NSW Government through the Department of Environment, Climate Change and Water. This document does not necessarily represent the opinions of the NSW Government or the Department of Environment, Climate Change and Water. Damon Telfer GECO Environmental 5 Arcadia Lane, GRASSY HEAD NSW 2441 Email: [email protected] © Damon Telfer, February 2010. This document is copyright and cannot be reproduced in part or whole without the express permission of the author. A permanent, irrevocable royalty-free, non-exclusive license to make these reports, documents and any other materials publicly available and to otherwise communicate, reproduce, adapt and publicise them on a non- profit basis is granted by the authors to Bellingen Shire Council and the Department of Environment, Climate Change and Water. -

The Moss-Back Alga (Cladophorophyceae, Chlorophyta) on Two Species of Freshwater Turtles in the Kimberleys
Telopea 12(2) 279–284 The moss-back alga (Cladophorophyceae, Chlorophyta) on two species of freshwater turtles in the Kimberleys Stephen Skinner1,2, Nancy FitzSimmons3 and Timothy J. Entwisle1 1National Herbarium of New South Wales, Mrs Macquaries Road, Sydney NSW 2000 Australia 2Southern ACT Catchment Group Inc., PO Box 2056, Kambah, ACT Author for correspondence: [email protected] 3Institute for Applied Ecology, School of Resource, Environmental & Heritage Sciences, University of Canberra, Canberra, ACT 2601, Australia Abstract The range of the Australian freshwater alga Basicladia ramulosa Ducker is extended, both in its turtle hosts (Chelodina burrungandjii Thomson et al.; Emydura australis (Grey)) and in geography, to tropical northern Western Australia. Along with further morphological observations, sporangia are described for the first time in this taxon. Introduction Moss-back turtles (Fig. 1) have fascinated biologists for many years. While the carapace of a potentially amphibious turtle would be a challenging habitat for most aquatic organisms, it is perhaps surprising there are only a handful of attached algae reported from such sites. Edgren et al. (1953) detailed the range of host turtles then known in North America and the range of epizoic algae that included Rhizoclonium and Cladophora. Two further genera in the Cladophoraceae are the only macroalgae widely reported on turtle carapaces: the prostrate, spreading, endozoic (and possibly disease causing) Dermatophyton radicans Peter, and species of the heterotrichous genus Basicladia, responsible for the name ‘moss-back’. In the United States, Basicladia is considered a small epizoic genus on turtles and water snails, of three to four taxa (John 2003). Hamilton (1948) described sexual reproduction in North American species of Basicladia involving the fusion of biflagellate zooids as is commonly the case in the Cladophoraceae. -

Eastern Snake-Necked Turtle
Husbandry Manual for Eastern Snake-Necked Turtle Chelodina longicollis Reptilia: Chelidae Image Courtesy of Jacki Salkeld Author: Brendan Mark Host Date of Preparation: 04/06/06 Western Sydney Institute of TAFE - Richmond Course Name and Number: 1068 Certificate 3 - Captive Animals Lecturers: Graeme Phipps/Andrew Titmuss/ Jacki Salkeld CONTENTS 1. Introduction 4 2. Taxonomy 5 2.1 Nomenclature 5 2.2 Subspecies 5 2.3 Synonyms 5 2.4 Other Common Names 5 3. Natural History 6 3.1 Morphometrics 6 3.1.1 Mass and Basic Body Measurements 6 3.1.2 Sexual Dimorphism 6 3.1.3 Distinguishing Features 7 3.2 Distribution and Habitat 7 3.3 Conservation Status 8 3.4 Diet in the Wild 8 3.5 Longevity 8 3.5.1 In the Wild 8 3.5.2 In Captivity 8 3.5.3 Techniques Used to Determine Age in Adults 9 4. Housing Requirements 10 4.1 Exhibit/Enclosure Design 10 4.2 Holding Area Design 10 4.3 Spatial Requirements 11 4.4 Position of Enclosures 11 4.5 Weather Protection 11 4.6 Temperature Requirements 12 4.7 Substrate 12 4.8 Nestboxes and/or Bedding Material 12 4.9 Enclosure Furnishings 12 5. General Husbandry 13 5.1 Hygiene and Cleaning 13 5.2 Record Keeping 13 5.3 Methods of Identification 13 5.4 Routine Data Collection 13 6. Feeding Requirements 14 6.1 Captive Diet 14 6.2 Supplements 15 6.3 Presentation of Food 15 1 7. Handling and Transport 16 7.1 Timing of Capture and Handling 16 7.2 Capture and Restraint Techniques 16 7.3 Weighing and Examination 17 7.4 Release 17 7.5 Transport Requirements 18 7.5.1 Box Design 18 7.5.2 Furnishings 19 7.5.3 Water and Food 19 7.5.4 Animals Per Box 19 7.5.5 Timing of Transportation 19 7.5.6 Release from Box 19 8. -

Testudines: Chelidae) of Australia, New Guinea and Indonesia
Zoological Journal of the Linnean Society, 2002, 134, 401–421. With 7 figures Electrophoretic delineation of species boundaries within the genus Chelodina (Testudines: Chelidae) of Australia, New Guinea and Indonesia ARTHUR GEORGES1*, MARK ADAMS2 and WILLIAM McCORD3 1Applied Ecology Research Group, University of Canberra, ACT 2601, Australia 2Evolutionary Biology Unit, South Australian Museum, North Terrace, Adelaide, SA 5001, Australia 3East Fishkill Animal Hospital, 285 Rt 82, Hopewell Junction NY 12533, USA Received February 2001; revised and accepted for publication June 2001 A total of 281 specimens of long-necked chelid turtles (Chelodina) were obtained from drainages of Australia, Papua New Guinea and the island of Roti in Indonesia. Ten diagnosable taxa were identified using allozyme profiles at 45 presumptive loci. Chelodina expansa, C. parkeri, C. rugosa and C. burrungandjii are in a Group A clade, C. longi- collis, C. novaeguineae, C. steindachneri, C. pritchardi and C. mccordi are in a Group B clade, and C. oblonga is in a monotypic Group C clade, with each clade thought to represent a distinct subgenus. Chelodina siebenrocki is syn- onymised with C. rugosa. An eleventh taxon, C. reimanni, could not be distinguished from C. novaeguineae on the basis of allozyme profiles, but it is morphologically distinct. Its status is therefore worthy of further investigation. Three instances of natural hybridization were detected. Chelodina rugosa and C. novaeguineae hybridize in the Gulf country of Queensland, with evidence of backcrossing to C. novaeguineae. Chelodina longicollis and C. novaeguineae hybridize in central coastal Queensland, and C. rugosa and C. burrungandjii hybridize along their zone of contact in the plateau escarpment streams and pools.