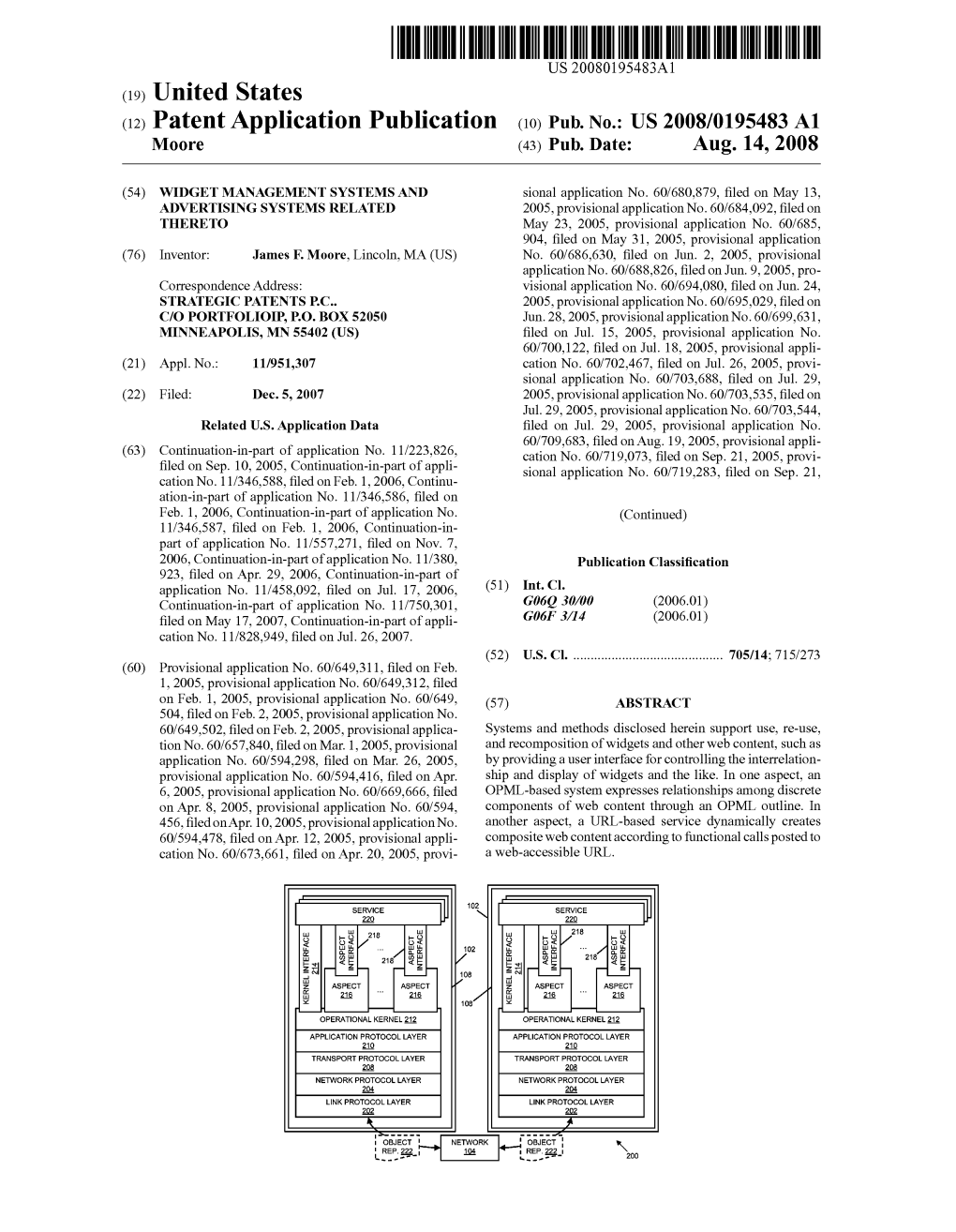
(12) Patent Application Publication (10) Pub. No.: US 2008/0195483 A1 M00re (43) Pub
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Glossary.Pdf
2 Contents 1 Glossary 4 3 1 Glossary Technologies Akonadi The data storage access mechanism for all PIM (Personal Information Manager) data in KDE SC 4. One single storage and retrieval system allows efficiency and extensibility not possible under KDE 3, where each PIM component had its own system. Note that use of Akonadi does not change data storage formats (vcard, iCalendar, mbox, maildir etc.) - it just provides a new way of accessing and updating the data.</p><p> The main reasons for design and development of Akonadi are of technical nature, e.g. having a unique way to ac- cess PIM-data (contacts, calendars, emails..) from different applications (e.g. KMail, KWord etc.), thus eliminating the need to write similar code here and there.</p><p> Another goal is to de-couple GUI applications like KMail from the direct access to external resources like mail-servers - which was a major reason for bug-reports/wishes with regard to perfor- mance/responsiveness in the past.</p><p> More info:</p><p> <a href=https://community.kde.org/KDE_PIM/Akonadi target=_top>Akonadi for KDE’s PIM</a></p><p> <a href=https://en.wikipedia.org/wiki/Akonadi target=_top>Wikipedia: Akonadi</a></p><p> <a href=https://techbase.kde.org/KDE_PIM/Akonadi target=_top>Techbase - Akonadi</a> See Also "GUI". See Also "KDE". Applications Applications are based on the core libraries projects by the KDE community, currently KDE Frameworks and previously KDE Platform.</p><p> More info:</p><p> <a href=https://community.kde.org/Promo/Guidance/Branding/Quick_Guide/ target=_top>KDE Branding</a> See Also "Plasma". -

Full Circle the INDEPENDENT MAGAZINE for the UBUNTU COMMUNITY
Issue #2 - June 2007 full circle THE INDEPENDENT MAGAZINE FOR THE UBUNTU COMMUNITY LINSPIRE & MICROSOFT LINSPIRE SIGNS FOR PATENT PROTECTION NEXT WHO'S REVIEW : HOW TO : INSTALL : SYSTEM76 LAPTOP UBUNTU ON A MAC MINI STEP-BY-STEP INSTALL FROM GETTING THE BOX BASIC INSTALL OF VPN OF KUBUNTU 7.04 : TO BOOTING UBUNTU UBUNTU FOR GRANDMA! FEISTY FAWN fullcircle magazine is not affiliated with or endorsed by Canonical Ltd. full circle News p.03 Your monthly update of what's going on in the world of Linux and Open Source Software Flavour of the Month - Kubuntu p.06 This month Robert Clipsham shows us a step-by-step STEP-BY-STEP INSTALL installation of Kubuntu Linux 7.04 P.06 How-To Ubuntu on the Mac Mini p.09 Ubuntu for Grandma p.12 Virtual Private Networking p.16 Scribus - Part 2 p.17 Review - System76 Laptop p.20 P.17 P.16 Thinking of buying a laptop? System76 supplies quality laptops pre-installed with Ubuntu! Interview - GRAMPS p.24 P12 P.20 My Story - 6 Months with Ubuntu p.29 Fighting to live with Linux after years of Windows use. Ubuntu Youth p.31 Letters p.33 Q&A p.35 My Desktop & My PC p.36 P.09 P.24 Show us your mean-machine! The Top 5 p.37 Andrew Min presents his Top5 Widgets How to Contribute p.39 Want to help fill future issues? Here are the details All text and images contained in this magazine are released under the Creative Commons Attribution-By-ShareAlike 3.0 Unported license. -

Desktop Paradise Had It with Human? Bored with Blue? Let Graham Morrison, Our Resident Aesthete, Guide You to Form and Function on the Linux Desktop
Get the perfect desktop Desktop paradise Had it with Human? Bored with blue? Let Graham Morrison, our resident aesthete, guide you to form and function on the Linux desktop. e all spend a great deal of time at the Linux the applications that let you change everything in the first place. desktop. It’s our office, living room and games Some changes are purely for decoration – a background image or room rolled into one. And like those rooms, the colour scheme, for instance. But there are others that reflect your Wexperience is a working routine, such as lot more pleasant when application icons in the toolbar you’ve made your “We’re going to cover and the keyboard shortcuts for environment more the areas where small launching them, and even the comfortable and more tuned font sizes and window position. to the way you work. It’s the changes yield big results.” Over the next few pages, virtual equivalent to moving we’re going to cover the main the sofa in front of the telly, putting beer in the fridge and areas on the desktop where small changes yield big results, and lighting a few candles. cover the main desktops most of us enjoy using. We hope you’ll be It might sound strange, but your desktop is an expression of inspired enough to make a few changes yourself. who you are, and how we use computers. That’s why the internet is littered with people who post images of their desktop, even going to the trouble of designing their own icon sets and building Under the microscope: Xfce KDE It’s often overlooked, but this is Forget blue, change the colours a desktop environment you can around and banish all thoughts truly make your mark on. -

Creating a Superkaramba Theme WIDGET BUILDER If You Can’T Find the Superkaramba Theme You’Re Looking For, You Can Also Lets You Always Build Your Own
KNOW-HOW SuperKaramba Workshop Creating a SuperKaramba theme WIDGET BUILDER www.sxc.hu If you can’t find the SuperKaramba theme you’re looking for, you can also lets you always build your own. BY HAGEN HÖPFNER specify if you want to keep the ability to move a theme. Some themes have their own configura- retty wallpaper is fine as far as it to get started with SuperKaramba. For tion dialogs. For example, you can con- goes, but if you want desktop additional information, see [2]. (These figure the weather report to match your Pfrills that actually do work for articles are currently available through current location. you, such as displaying weather reports Linux Magazine’s convenient online ar- or monitoring system data, you’ll need a chive.) This article is a brief workshop Creating Your Own Themes tool like SuperKaramba. SuperKaramba on how to create your own Super- To create your own theme, you need to [1] is a KDE-based tool that lets you cre- Karamba theme. create a file, say my_theme.theme, which ate helpful widgets for your desktop. you can open in SuperKaramba as a The SuperKaramba website refers to Finding the Software local file. As graphics and Python scripts these interactive widgets as “custom eye The current SuperKaramba version is can be added to the layout file, you will candy,” but SuperKaramba applets are number 0.37. If you prefer not to build probably want to store all of these com- more commonly called themes. You’ll SuperKaramba yourself, you will find ponents in a single directory. -

Download an Upgrade
ALAGAPPA UNIVERSITY (Accredited with ‘A+’ Grade by NAAC (with CGPA: 3.64) in the Third Cycle and Graded as category - I University by MHRD-UGC) (A State University Established by the Government of Tamilnadu) KARAIKUDI – 630 003 DIRECTORATE OF DISTANCE EDUCATION M.Sc. (Computer Science) Second Year – Third Semester 341 32 CLOUD COMPUTING Copy Right Reserved For Private Use only i Author: Dr. C.Balakrishnan Assistant Pro fessor Alagappa Institute of Skill Development Alagappa University, Karaikudi. 630 003. “The Copyright shall be vested with Alagappa University” All rights reserved. No part of this publication which is material protected by this copyright notice may be reproduced or transmitted or utilized or stored in any form or by any means now known or hereinafter invented, electronic, digital or mechanical, including photocopying, scanning, recording or by any information storage or retrieval system, without prior written permission from the Alagappa University, Karaikudi, Tamil Nadu. Reviewer: Mr. S. Balasubramanian Assistant Professor in Computer Science Directorate of Distance Education Alagappa University Karaikudi – 03. ii CLOUD COMPUTING SYLLABI-BOOK MAPPING TABLE Syllabi Mapping in Book UNIT BLOCK 1 : CLOUD COMPUTING BASICS 1-23 Introduction: History – Working with cloud computing – pros and 1 1-11 cons of cloud computing – Benefits Developing cloud services – pros and cons of cloud service 2 12-17 development - types of cloud service development 3 Discovering cloud services development services and tools 18-23 BLOCK 2 : CLOUD -

What Is Cloud Computing? Cloud Computing Refers to Manipulating, Configuring, and Accessing the Applications Online
What is Cloud? The term Cloud refers to a Network or Internet. In other words, we can say that Cloud is something, which is present at remote location. Cloud can provide services over network, i.e., on public networks or on private networks, i.e., WAN, LAN or VPN. Applications such as e-mail, web conferencing, customer relationship management (CRM),all run in cloud. What is Cloud Computing? Cloud Computing refers to manipulating, configuring, and accessing the applications online. It offers online data storage, infrastructure and application. We need not to install a piece of software on our local PC and this is how the cloud computing overcomes platform dependency issues. Hence, the Cloud Computing is making our business application mobile and collaborative. Basic Concepts There are certain services and models working behind the scene making the cloud computing feasible and accessible to end users. Following are the working models for cloud computing: • Deployment Models • Service Models DEPLOYMENT MODELS Deployment models define the type of access to the cloud, i.e., how the cloud is located? Cloud can have any of the four types of access: Public, Private, Hybrid and Community. PUBLIC CLOUD The Public Cloud allows systems and services to be easily accessible to the general public. Public cloud may be less secure because of its openness, e.g., e-mail. PRIVATE CLOUD The Private Cloud allows systems and services to be accessible within an organization. It offers increased security because of its private nature. COMMUNITY CLOUD The Community Cloud allows systems and services to be accessible by group of organizations. -
Free and Open Source Software
Free and open source software Copyleft ·Events and Awards ·Free software ·Free Software Definition ·Gratis versus General Libre ·List of free and open source software packages ·Open-source software Operating system AROS ·BSD ·Darwin ·FreeDOS ·GNU ·Haiku ·Inferno ·Linux ·Mach ·MINIX ·OpenSolaris ·Sym families bian ·Plan 9 ·ReactOS Eclipse ·Free Development Pascal ·GCC ·Java ·LLVM ·Lua ·NetBeans ·Open64 ·Perl ·PHP ·Python ·ROSE ·Ruby ·Tcl History GNU ·Haiku ·Linux ·Mozilla (Application Suite ·Firefox ·Thunderbird ) Apache Software Foundation ·Blender Foundation ·Eclipse Foundation ·freedesktop.org ·Free Software Foundation (Europe ·India ·Latin America ) ·FSMI ·GNOME Foundation ·GNU Project ·Google Code ·KDE e.V. ·Linux Organizations Foundation ·Mozilla Foundation ·Open Source Geospatial Foundation ·Open Source Initiative ·SourceForge ·Symbian Foundation ·Xiph.Org Foundation ·XMPP Standards Foundation ·X.Org Foundation Apache ·Artistic ·BSD ·GNU GPL ·GNU LGPL ·ISC ·MIT ·MPL ·Ms-PL/RL ·zlib ·FSF approved Licences licenses License standards Open Source Definition ·The Free Software Definition ·Debian Free Software Guidelines Binary blob ·Digital rights management ·Graphics hardware compatibility ·License proliferation ·Mozilla software rebranding ·Proprietary software ·SCO-Linux Challenges controversies ·Security ·Software patents ·Hardware restrictions ·Trusted Computing ·Viral license Alternative terms ·Community ·Linux distribution ·Forking ·Movement ·Microsoft Open Other topics Specification Promise ·Revolution OS ·Comparison with closed -

Downloaded from Public Repositories Such As Docker Hub
bioRxiv preprint doi: https://doi.org/10.1101/099010; this version posted November 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Hung et al. Building containerized workflows using the BioDepot-workflow-builder (Bwb) Ling-Hong Hung1, Jiaming Hu1, Trevor Meiss1, Alyssa Ingersoll1, Wes Lloyd1, Daniel Kristiyanto1, Yuguang Xiong2, Eric Sobie2 and Ka Yee Yeung1* *Correspondence: [email protected] 1School of Engineering and Abstract Technology, University of Washington, Box 358426, 98402 We present the BioDepot-workflow-builder (Bwb), a software tool that allows Tacoma WA, USA users to create and execute reproducible bioinformatics workflows using a Full list of author information is drag-and-drop interface. Graphical widgets represent Docker containers executing available at the end of the article a modular task. Widgets are then graphically linked to build bioinformatics workflows that can be reproducibly deployed across different local and cloud platforms. Each widget contains a form-based user interface to facilitate parameter entry and a console to display intermediate results. Bwb provides tools for rapid customization of widgets, containers and workflows. Saved workflows can be shared using Bwb's native format or exported as shell scripts. Keywords: reproducibility of research; bioinformatics workflows; software development; RNA-seq; Docker Background One of the key challenges for biomedical science is the rapidly increasing number and complexity of analytical methods. Reproducing the results of a bioinformat- ics workflow can be challenging given the number of components, each of which having its own set of parameters, dependencies, supporting files, and installation requirements. -

UCI-AM-17-041.Pdf
UCI-AM-17-041 ACQUISITION RESEARCH PROGRAM SPONSORED REPORT SERIES Achieving Better Buying Power for Mobile Open Architecture Software Systems through Diverse Acquisition Scenarios 1 May 2017 Dr. Walt Scacchi Dr. Thomas A. Alspaugh Institute for Software Research University of California, Irvine Disclaimer: This material is based upon work supported by the Naval Postgraduate School Acquisition Research Program under Grant No. N00244-16-1-0004. The views expressed in written materials or publications, and/or made by speakers, moderators, and presenters, do not necessarily reflect the official policies of the Naval Postgraduate School nor does mention of trade names, commercial practices, or organizations imply endorsement by the U.S. Government. Approved for public release; distribution is unlimited. Prepared for the Naval Postgraduate School, Monterey, CA 93943. Acquisition Research Program Graduate School of Business & Public Policy Naval Postgraduate School The research presented in this report was supported by the Acquisition Research Program of the Graduate School of Business & Public Policy at the Naval Postgraduate School. To request defense acquisition research, to become a research sponsor, or to print additional copies of reports, please contact any of the staff listed on the Acquisition Research Program website (www.acquisitionresearch.net). Acquisition Research Program Graduate School of Business & Public Policy Naval Postgraduate School Executive Summary This research seeks to identify, track, and analyze software component costs and cost reduction opportunities within diverse acquisition life cycle scenarios for open architecture systems accommodating Web-based and mobile devices, where such systems combine best-of- breed software components and software products lines subject to different intellectual property license and cybersecurity requirements. -
(12) United States Patent (10) Patent No.: US 7.793,232 B2 Chaudhri Et Al
US007793232B2 (12) United States Patent (10) Patent No.: US 7.793,232 B2 Chaudhri et al. (45) Date of Patent: Sep. 7, 2010 (54) UNIFIED INTEREST LAYER FOR USER 5,289,574 A 2/1994 Sawyer INTERFACE (75) Inventors: Imran A. Chaudhri, San Francisco, CA (US); John Louch, San Luis Obispo, CA (Continued) (US); Andrew M. Grignon, Campbell, CA (US); Gregory N. Christie, San FOREIGN PATENT DOCUMENTS Jose, CA (US) EP 548586 A2 6, 1993 (73) Assignee: Apple Inc., Cupertino, CA (US) (*) Notice: Subject to any disclaimer, the term of this patent is extended or adjusted under 35 (Continued) U.S.C. 154(b) by 674 days. OTHER PUBLICATIONS (21) Appl. No.: 11/370,743 Microsoft et al. “Microsoft Windows XP Microsoft Word 2003, 2003.* (22) Filed: Mar. 7, 2006 (Continued) (65) Prior Publication Data Primary Examiner Steven BTheriault US 2006/O15O118A1 Jul. 6, 2006 (74) Attorney, Agent, or Firm Fenwick & West LLP Related U.S. Application Data (57) ABSTRACT (63) Continuation of application No. 10/877.968, filed on Jun. 25, 2004. A user-activatable dashboard (also referred to as a unified (51) Int. C. interest layer) contains any number of user interface ele G6F 3/48 (2006.01) ments, referred to herein as “widgets.” for quick access by a GO6F 3/OO (2006.01) user. In response to a command from a user, the dashboard is (52) U.S. Cl. ....................... 715/802; 709/217; 709/218; invoked and the widgets are shown on the screen. The user can 709/219; 715/762; 715/763; 715/764; 715/765; activate the dashboard at any time, causing the dashboard to 715/767; 715/781: 715/782; 715/792; 715/840; temporarily replace the existing user interface display on the 71.5/841 user's screen. -

D5.4.1-Integrated Dicode Services V5 EC
Mastering Data-Intensive Collaboration and Decision Making FP7 - Information and Communication Technologies Grant Agreement no: 257184 Collaborative Project Project start: 1 September 2010, Duration: 36 months D5.4.1 – Integrated Dicode services (initial version) Due date of deliverable: 30 November 2011 Actual submission date: 30 November 2011 Responsible Partner: UPM Contributing Partners: UPM, CTI, UOL, BRF Nature: Report Prototype 3 Demonstrator 3 Other Dissemination Level: PU : Public PP : Restricted to other programme participants (including the Commission Services) RE : Restricted to a group specified by the consortium (including the Commission Services) CO : Confidential, only for members of the consortium (including the Commission Services) Keyword List: Service Integration, Integration Framework, Integrated Workbench, User Interface, Web Application, Widgets, Service Oriented Architecture The Dicode project (http://dicode-project.eu/) is funded by the European Commission, Information Society and Media Directorate General, under the FP7 Cooperation programme (ICT/SO 4.3: Intelligent Information Management). FP7-ICT-2009-5 257184 - Dicode The Dicode Consortium Computer Technology Institute & Press “Diophantus” (CTI) (coordinator), Greece University of Leeds (UOL), UK Fraunhofer-Gesellschaft zur Foerderung der angewandten Forschung e.V. (FHG), Germany Universidad Politécnica de Madrid (UPM), Spain Neofonie GmbH (NEO), Germany Image Analysis Limited (IMA), UK Biomedical Research Foundation, Academy of Athens (BRF), Greece -

Embedded Business Intelligence Market Study
November 30, 2017 Dresner Advisory Services, LLC 2017 Edition Embedded Business Intelligence Market Study Wisdom of Crowds® Series Licensed to MicroStrategy 2017 Embedded Business Intelligence Market Study Disclaimer: This report should be used for informational purposes only. Vendor and product selections should be made based on multiple information sources, face-to-face meetings, customer reference checking, product demonstrations, and proof-of-concept applications. The information contained in all Wisdom of Crowds® Market Study Reports reflects the opinions expressed in the online responses of individuals who chose to respond to our online questionnaire and does not represent a scientific sampling of any kind. Dresner Advisory Services, LLC shall not be liable for the content of reports, study results, or for any damages incurred or alleged to be incurred by any of the companies included in the reports as a result of its content. Reproduction and distribution of this publication in any form without prior written permission is forbidden. http://www.dresneradvisory.com Copyright 2017 – Dresner Advisory Services, LLC 2 2017 Embedded Business Intelligence Market Study Definitions Business Intelligence Defined Business intelligence (BI) is “knowledge gained through the access and analysis of business information. Business Intelligence tools and technologies include query and reporting, OLAP (on-line analytical processing), data mining and advanced analytics, end-user tools for ad hoc query and analysis,” and “dashboards for performance monitoring.” Howard Dresner, The Performance Management Revolution: Business Results Through Insight and Action (John Wiley & Sons, 2007) Embedded Business Intelligence Defined Embedded business intelligence is the technological capability to include BI features and functions as an inherent part of another application.