Reactome | Metabolism of Amino Acids and Derivatives (R-HSA-71291)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PROTEOMIC ANALYSIS of HUMAN URINARY EXOSOMES. Patricia
ABSTRACT Title of Document: PROTEOMIC ANALYSIS OF HUMAN URINARY EXOSOMES. Patricia Amalia Gonzales Mancilla, Ph.D., 2009 Directed By: Associate Professor Nam Sun Wang, Department of Chemical and Biomolecular Engineering Exosomes originate as the internal vesicles of multivesicular bodies (MVBs) in cells. These small vesicles (40-100 nm) have been shown to be secreted by most cell types throughout the body. In the kidney, urinary exosomes are released to the urine by fusion of the outer membrane of the MVBs with the apical plasma membrane of renal tubular epithelia. Exosomes contain apical membrane and cytosolic proteins and can be isolated using differential centrifugation. The analysis of urinary exosomes provides a non- invasive means of acquiring information about the physiological or pathophysiological state of renal cells. The overall objective of this research was to develop methods and knowledge infrastructure for urinary proteomics. We proposed to conduct a proteomic analysis of human urinary exosomes. The first objective was to profile the proteome of human urinary exosomes using liquid chromatography-tandem spectrometry (LC- MS/MS) and specialized software for identification of peptide sequences from fragmentation spectra. We unambiguously identified 1132 proteins. In addition, the phosphoproteome of human urinary exosomes was profiled using the neutral loss scanning acquisition mode of LC-MS/MS. The phosphoproteomic profiling identified 19 phosphorylation sites corresponding to 14 phosphoproteins. The second objective was to analyze urinary exosomes samples isolated from patients with genetic mutations. Polyclonal antibodies were generated to recognize epitopes on the gene products of these genetic mutations, NKCC2 and MRP4. The potential usefulness of urinary exosome analysis was demonstrated using the well-defined renal tubulopathy, Bartter syndrome type I and using the single nucleotide polymorphism in the ABCC4 gene. -

Glycine and Serine Inhibition of D-Glycerate Dehydrogenase and 3-Phosphoglycerate Dehydrogenase of Rat Brain
Volume 17, number 1 FEBS LETTERS September 1971 GLYCINE AND SERINE INHIBITION OF D-GLYCERATE DEHYDROGENASE AND 3-PHOSPHOGLYCERATE DEHYDROGENASE OF RAT BRAIN M.L. UHR and M.K. SNEDDON Department of Physiology, Australian National University, Canberra, A.C. T. 2601, Australia Received 16 July 1971 1. Introduction 2. Materials and methods Glycine is probably a major inhibitory transmitter Barium phosphoglycerate and calcium DL-glycer- in the mammalian central nervous system [ 1,2] . ate purchased from Sigma Chemical Corporation, were Hence the metabolism and metabolic control of converted to sodium salts by passage through a glycine could be important for the efficient function- column of Dowex-SO(H?) and neutralization of the ing of nervous tissue, particularly of the spinal cord. emerging acids. Phosphoglycerate concentration was As glycine may be formed from serine by serine estimated by the method of Czok and Eckert [7]. hydroxymethylase (EC 2.1.2.1) [3] , enzymes neces- Glycerate was estimated by the method of Bartlett sary for the formation of serine from carbohydrate [8] with the concentration of chromotrophic acid sources could be important for the production and raised to 0.025% as recommended by Dawkins and regulation of glycine. Two major pathways of serine Dickens [9] . NAD was purchased from P-L Biochemi- formation have been described in mammalian sys- cals, and NADP from Sigma. tems, the “phosphoryl:ted” pathway from 3-phos- Rat cortical tissue was homogenized in 0.32 M phoglycerate [4], and the “non-phosphorylated” sucrose containing 0.5 mM dithiothreitol (DTT) and pathway from D-glycerate [5,6]. We decided there- centrifuged at 105,000 g for 100 min. -

Severe Child Form of Primary Hyperoxaluria Type 2
Konkoľová et al. BMC Medical Genetics (2017) 18:59 DOI 10.1186/s12881-017-0421-8 CASE REPORT Open Access Severe child form of primary hyperoxaluria type 2 - a case report revealing consequence of GRHPR deficiency on metabolism Jana Konkoľová1,2*, Ján Chandoga1,2, Juraj Kováčik3, Marcel Repiský2, Veronika Kramarová2, Ivana Paučinová3 and Daniel Böhmer1,2 Abstract Background: Primary hyperoxaluria type 2 is a rare monogenic disorder inherited in an autosomal recessive pattern. It results from the absence of the enzyme glyoxylate reductase/hydroxypyruvate reductase (GRHPR). As a consequence of deficient enzyme activity, excessive amounts of oxalate and L-glycerate are excreted in the urine, and are a source for the formation of calcium oxalate stones that result in recurrent nephrolithiasis and less frequently nephrocalcinosis. Case presentation: We report a case of a 10-month-old patient diagnosed with urolithiasis. Screening of inborn errors of metabolism, including the performance of GC/MS urine organic acid profiling and HPLC amino acid profiling, showed abnormalities, which suggested deficiency of GRHPR enzyme. Additional metabolic disturbances observed in the patient led us to seek other genetic determinants and the elucidation of these findings. Besides the elevated excretion of 3-OH-butyrate, adipic acid, which are typical marks of ketosis, other metabolites such as 3- aminoisobutyric acid, 3-hydroxyisobutyric acid, 3-hydroxypropionic acid and 2-ethyl-3-hydroxypropionic acids were observed in increased amounts in the urine. Direct sequencing of the GRHPR gene revealed novel mutation, described for the first time in this article c.454dup (p.Thr152Asnfs*39) in homozygous form. The frequent nucleotide variants were found in AGXT2 gene. -

In Silico Prediction of the Mode of Action of Viola Odorata in Diabetes
Hindawi BioMed Research International Volume 2020, Article ID 2768403, 13 pages https://doi.org/10.1155/2020/2768403 Research Article In Silico Prediction of the Mode of Action of Viola odorata in Diabetes Manal Ali Buabeid ,1 El-Shaimaa A. Arafa ,1,2 Waseem Hassan ,3 and Ghulam Murtaza 3 1College of Pharmacy and Health Sciences, Ajman University, Ajman 346, UAE 2Department of Pharmacology and Toxicology, Faculty of Pharmacy, Beni-Suef University, Beni Suef 62514, Egypt 3Department of Pharmacy, COMSATS University Islamabad, Lahore Campus, 54000, Pakistan Correspondence should be addressed to Manal Ali Buabeid; [email protected] Received 16 April 2020; Revised 27 June 2020; Accepted 5 October 2020; Published 31 October 2020 Academic Editor: K. H. Mok Copyright © 2020 Manal Ali Buabeid et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. The metabolic syndrome increases the risk of different diseases such as type 2 diabetes. The prevalence of metabolic syndrome has rapidly grown and affected more than 230 million people worldwide. Viola odorata is a traditionally used plant for the treatment of diabetes; however, its mechanism to manage diabetes is still unknown. Purpose. This study was designed to systematically assess the mechanism of action of Viola odorata in diabetes. Methods. An extensive literature search was made to establish an ingredient-target database of Viola odorata. Of these, targets related to diabetes were identified and used to develop a protein-protein interaction network (PPIN) by utilizing the STITCH database. -

Dependent Malic Enzyme: a New Class of Oxidative Decarboxylases Yingwu Xu1, Girija Bhargava1, Hao Wu2, Gerhard Loeber3 and Liang Tong1*
Research Article 877 Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases Yingwu Xu1, Girija Bhargava1, Hao Wu2, Gerhard Loeber3 and Liang Tong1* Background: Malic enzymes catalyze the oxidative decarboxylation of malate to Addresses: 1Department of Biological Sciences, pyruvate and CO with the concomitant reduction of NAD(P)+ to NAD(P)H. Columbia University, New York, NY 10027, USA, 2 2Department of Biochemistry, The Weill Medical They are widely distributed in nature and have important biological functions. College of Cornell University, New York, NY 10021, Human mitochondrial NAD(P)+-dependent malic enzyme (mNAD-ME) may have USA and 3Boehringer Ingelheim Austria R&D, Dr a crucial role in the metabolism of glutamine for energy production in rapidly Boehringer-Gasse 5-11, A-1121 Vienna, Austria. dividing cells and tumors. Moreover, this isoform is unique among malic enzymes *Corresponding author. in that it is a cooperative enzyme, and its activity is controlled allosterically. E-mail: [email protected] Results: The crystal structure of human mNAD-ME has been determined at Keywords: allosteric interactions, cofactor 2.5 Å resolution by the selenomethionyl multiwavelength anomalous diffraction specificity, glutamine metabolism, protein folds method and refined to 2.1 Å resolution. The structure of the monomer can be Received: 11 March 1999 divided into four domains; the active site of the enzyme is located in a deep Revisions requested: 7 April 1999 cleft at the interface between three of the domains. Three acidic residues Revisions received: 14 April 1999 (Glu255, Asp256 and Asp279) were identified as ligands for the divalent cation Accepted: 15 April 1999 that is required for catalysis by malic enzymes. -
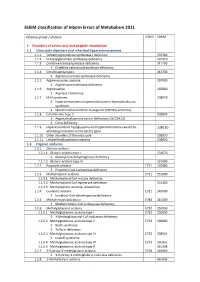
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

An Archaeal ADP-Dependent Serine Kinase Involved in Cysteine Biosynthesis and Serine Metabolism
ARTICLE Received 4 Feb 2016 | Accepted 5 Oct 2016 | Published 18 Nov 2016 DOI: 10.1038/ncomms13446 OPEN An archaeal ADP-dependent serine kinase involved in cysteine biosynthesis and serine metabolism Yuki Makino1, Takaaki Sato1,2, Hiroki Kawamura1, Shin-ichi Hachisuka1,2, Ryo Takeno1, Tadayuki Imanaka2,3 & Haruyuki Atomi1,2 Routes for cysteine biosynthesis are still unknown in many archaea. Here we find that the hyperthermophilic archaeon Thermococcus kodakarensis generates cysteine from serine via O-phosphoserine, in addition to the classical route from 3-phosphoglycerate. The protein responsible for serine phosphorylation is encoded by TK0378, annotated as a chromosome partitioning protein ParB. The TK0378 protein utilizes ADP as the phosphate donor, but in contrast to previously reported ADP-dependent kinases, recognizes a non-sugar substrate. Activity is specific towards free serine, and not observed with threonine, homoserine and serine residues within a peptide. Genetic analyses suggest that TK0378 is involved in serine assimilation and clearly responsible for cysteine biosynthesis from serine. TK0378 homologs, present in Thermococcales and Desulfurococcales, are most likely not ParB proteins and constitute a group of kinases involved in serine utilization. 1 Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto 615-8510, Japan. 2 JST, CREST, 7, Gobancho, Chiyoda-ku, Tokyo 102-0076, Japan. 3 Research Organization of Science and Technology, Ritsumeikan University, Noji-Higashi, Kusatsu 525-8577, Japan. Correspondence and requests for materials should be addressed to H.A. (email: [email protected]). NATURE COMMUNICATIONS | 7:13446 | DOI: 10.1038/ncomms13446 | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/ncomms13446 embers of the Archaea, which form the third domain Evaluation of established Cys biosynthesis pathways. -

1 Hyperactive Mtor Induces Neuroendocrine Differentiation in Prostate Cancer Cell
1 Hyperactive mTOR Induces Neuroendocrine Differentiation in Prostate Cancer Cell 2 with Concurrent Up-regulation of IRF1 3 4 Authors: Mayuko Kanayama1, 2, Toshiya Hayano3, Michinori Koebis2, Tatsuya Maeda4, Yoko 5 Tabe5, Shigeo Horie1, and Atsu Aiba2 6 7 Affiliations: 1Department of Urology, Juntendo University Graduate School of Medicine, 8 Tokyo, Japan, 2Laboratory of Animal Resources, Center for Disease Biology and Integrated 9 Medicine, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan, 10 3Department of Biomedical Sciences, College of Life Sciences, Ritsumeikan University, 11 Shiga, Japan, 4Institute of Molecular and Cellular Biosciences, The University of Tokyo, 12 Tokyo, Japan, 5Department of Clinical Laboratory Medicine, Juntendo University Graduate 13 School of Medicine, Tokyo, Japan. 14 Correspondence: Atsu Aiba, Laboratory of Animal Resources, Center for Disease Biology 15 and Integrated Medicine, Graduate School of Medicine, The University of Tokyo, 7-3-1 16 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. Tel: +81-3-5841-3638, Fax: +81-3-5841-3679, 17 E-mail: [email protected] 18 The running title: The role of IRF1 in active mTOR-induced NED 19 This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative 20 Areas (Comprehensive Brain Science Network), Grant Numbers 221S0003 (to A.A.), and 21 Grant-in-Aid for Scientific Research (B), JSPS KAKENHI Grant Numbers 25291042 and 22 17H03802 (to T.M.) from the Ministry of Education, Science, Sports and Culture of Japan. 23 Disclosure of Potential Conflicts of Interest: The authors have no conflict of interest. 1 1 Abstract 2 BACKGROUND 3 Neuroendocrine-differentiated prostate cancer (NEPCa) is refractory to androgen deprivation 4 therapy and shows a poor prognosis. -

Kinetics and Regulation of the NAD(P)H-Dependent Glyoxylate-Specific Reductase from Spinach Leaves Leszek A
Kinetics and Regulation of the NAD(P)H-Dependent Glyoxylate-Specific Reductase from Spinach Leaves Leszek A. Kleczkowski Plant Physiology Department, University of Urnea, 901-87 Urnea, Sweden Z. Naturforsch. 50c, 21-28 (1995); received October 21/November 21, 1994 Alternative Substrate, Cyanide, Cytosol, Glycolate Pathway, Glyoxylate Reductase Kinetic mechanism of purified spinach leaf NAD(P)H glyoxylate reductase (GR-1) was studied using either NADPH and NADH as alternative substrates with glyoxylate. The mech anism was elucidated from substrate kinetic patterns using NADH as a cofactor rather than NADPH. With NADPH varied versus glyoxylate, and with NADPH and glyoxylate varied at a constant ratio, the patterns obtained on double reciprocal plots appeared to be consistent with a ping-pong mechanism; however, kinetic patterns with NADH conclusively ruled out the ping-pong reaction in favour of the sequential addition of the reactants. Product inhi bition studies with glycolate and NADP have suggested either that NADPH binds to the enzyme before glyoxylate or that the addition of substrates is a random one. Studies with active group modifiers suggested an involvement of histidine, serine and cysteine residues in GR-1 activity. Salts had little or no effect on the activity of the enzyme, with the exception of cyanide, which had an apparent K, of ca. 2 m M . Studies with several metabolites used as possible effectors of GR-1 activity have suggested that the enzyme is modulated only by substrate availability in vivo. The apparent insensitivity of GR-1 to metabolic effectors is consistent with the proposed role of the enzyme in detoxifying glyoxylate which may act as a potent inhibitor of photosynthetic processes in plant tissues. -

Short Abstinence: Impact on the Seminal Plasma Proteome and Accessory Sex Gland Secretions Thesis Presented in Fulfilment Of
Short abstinence: Impact on the seminal plasma proteome and accessory sex gland secretions By Dale Mark Goss Thesis presented in fulfilment of the requirements for the degree of Master of Science in the Faculty of Medicine and Health Sciences at Stellenbosch University Supervisor: Professor Stefan S. Du Plessis Co-supervisor: Professor Gerhard van der Horst Stellenbosch University https://scholar.sun.ac.za Declaration By submitting this dissertation, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof, that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. Dale M. Goss March 2018 Copyright © 2018 Stellenbosch University All rights reserved i Stellenbosch University https://scholar.sun.ac.za Abstract Various studies have sought to determine the typical versus optimal abstinence period after which semen samples should be collected, and many have been found to be contradictory. Several factors influence the semen microenvironment and subsequent sperm basic and functional parameters. In this study we focused on important biomarkers of prostate, seminal vesicle and epididymal secretion. Furthermore, extensive proteomic analysis of seminal plasma was performed for a more comprehensive understanding of the observations. Semen samples were obtained from normozoospermic donors (n = 16) after 4 days and 4 hours of ejaculatory abstinence (EA) and standard semen analysis was performed with the assistance of computer aided sperm analysis (CASA), while the seminal plasma citric acid, neutral alpha- glucosidase and fructose concentrations were measured photometrically with commercially available assay kits. -

Inhibitory Cross-Talk Upon Introduction of a New Metabolic Pathway Into an Existing Metabolic Network
Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network Juhan Kim and Shelley D. Copley1 Department of Molecular, Cellular, and Developmental Biology and Cooperative Institute for Research in Environmental Sciences, University of Colorado, Boulder, CO 80309 Edited by Gregory A. Petsko, Brandeis University, Waltham, MA, and approved August 2, 2012 (received for review May 21, 2012) Evolution or engineering of novel metabolic pathways can endow ample, pathways for degradation of pesticides such as atrazine microbes with new abilities to degrade anthropogenic pollutants and pentachlorophenol likely arose due to selective pressure for or synthesize valuable chemicals. Most studies of the evolution of use of a novel source of nitrogen (atrazine) or for elimination of new pathways have focused on the origins and quality of function a toxin as well as utilization of a new source of carbon (penta- of the enzymes involved. However, there is an additional layer of chlorophenol) (3). Evolution of strains capable of efficient bio- complexity that has received less attention. Introduction of a novel degradation is likely to require not only recruitment of the enzymes pathway into an existing metabolic network can result in inhibitory necessary to degrade a novel compound but also adaptations that cross-talk due to adventitious interactions between metabolites prevent new metabolites from interfering with the functions of and macromolecules that have not previously encountered one preexisting enzymes and transcriptional regulators. another. Here, we report a thorough examination of inhibitory Adventitious interactions between novel compounds and cross-talk between a novel metabolic pathway for synthesis of proteins in microbial metabolic networks also pose significant pyridoxal 5′-phosphate and the existing metabolic network of problems for efforts to engineer microbes for environmentally Escherichia coli. -

Pathway Search Result
Pathway Search Result Following object(s) was/were not found ec:2.7.11.24 ec:3.1.3.16 Sort by the pathway list Show all objects ec01100 Metabolic pathways (168) ec:1.1.1.100 3-oxoacyl-[acyl-carrier-protein] reductase; beta-ketoacyl-[acyl-carrier protein](ACP) reductase; beta-ketoacyl acyl carrier protein (ACP) reductase; beta-ketoacyl reductase; beta-ketoacyl thioester reductase; beta-ketoacyl-ACP reductase; beta-ketoacyl-acyl carrier protein reductase; 3-ketoacyl acyl carrier protein reductase; NADPH-specific 3-oxoacyl-[acylcarrier protein]reductase; 3-oxoacyl-[ACP]reductase; (3R)-3-hydroxyacyl-[acyl- ec:1.1.1.205 IMP dehydrogenase; inosine-5'-phosphate dehydrogenase; inosinic acid dehydrogenase; inosinate dehydrogenase; inosine 5'-monophosphate dehydrogenase; inosine monophosphate dehydrogenase; IMP oxidoreductase; inosine monophosphate oxidoreductase ec:1.1.1.22 UDP-glucose 6-dehydrogenase; UDP-glucose dehydrogenase; uridine diphosphoglucose dehydrogenase; UDPG dehydrogenase; UDPG:NAD oxidoreductase; UDP-alpha-D-glucose:NAD oxidoreductase; UDP-glucose:NAD+ oxidoreductase; uridine diphosphate glucose dehydrogenase; UDP-D-glucose dehydrogenase; uridine diphosphate D-glucose dehydrogenase ec:1.1.1.267 1-deoxy-D-xylulose-5-phosphate reductoisomerase; DXP-reductoisomerase; 1-deoxy-D-xylulose-5-phosphate isomeroreductase; 2-C-methyl-D-erythritol 4-phosphate (MEP) synthase ec:1.1.1.29 glycerate dehydrogenase; D-glycerate dehydrogenase; hydroxypyruvate reductase; (R)-glycerate:NAD+ oxidoreductase ec:1.1.1.294 chlorophyll(ide) b reductase; chlorophyll