Cercopithecus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Targets, Tactics, and Cooperation in the Play Fighting of Two Genera of Old World Monkeys (Mandrillus and Papio): Accounting for Similarities and Differences
2019, 32 Heather M. Hill Editor Peer-reviewed Targets, Tactics, and Cooperation in the Play Fighting of Two Genera of Old World Monkeys (Mandrillus and Papio): Accounting for Similarities and Differences Kelly L. Kraus1, Vivien C. Pellis1, and Sergio M. Pellis1 1 Department of Neuroscience, University of Lethbridge, Canada Play fighting in many species involves partners competing to bite one another while avoiding being bitten. Species can differ in the body targets that are bitten and the tactics used to attack and defend those targets. However, even closely related species that attack and defend the same body target using the same tactics can differ markedly in how much the competitiveness of such interactions is mitigated by cooperation. A degree of cooperation is necessary to ensure that some turn-taking between the roles of attacker and defender occurs, as this is critical in preventing play fighting from escalating into serious fighting. In the present study, the dyadic play fighting of captive troops of 4 closely related species of Old World monkeys, 2 each from 2 genera of Papio and Mandrillus, was analyzed. All 4 species have a comparable social organization, are large bodied with considerable sexual dimorphism, and are mostly terrestrial. In all species, the target of biting is the same – the area encompassing the upper arm, shoulder, and side of the neck – and they have the same tactics of attack and defense. However, the Papio species exhibit more cooperation in their play than do the Mandrillus species, with the former using tactics that make biting easier to attain and that facilitate close bodily contact. -

Body Measurements for the Monkeys of Bioko Island, Equatorial Guinea
Primate Conservation 2009 (24): 99–105 Body Measurements for the Monkeys of Bioko Island, Equatorial Guinea Thomas M. Butynski¹,², Yvonne A. de Jong² and Gail W. Hearn¹ ¹Bioko Biodiversity Protection Program, Drexel University, Philadelphia, PA, USA ²Eastern Africa Primate Diversity and Conservation Program, Nanyuki, Kenya Abstract: Bioko Island, Equatorial Guinea, has a rich (eight genera, 11 species), unique (seven endemic subspecies), and threat- ened (five species) primate fauna, but the taxonomic status of most forms is not clear. This uncertainty is a serious problem for the setting of priorities for the conservation of Bioko’s (and the region’s) primates. Some of the questions related to the taxonomic status of Bioko’s primates can be resolved through the statistical comparison of data on their body measurements with those of their counterparts on the African mainland. Data for such comparisons are, however, lacking. This note presents the first large set of body measurement data for each of the seven species of monkeys endemic to Bioko; means, ranges, standard deviations and sample sizes for seven body measurements. These 49 data sets derive from 544 fresh adult specimens (235 adult males and 309 adult females) collected by shotgun hunters for sale in the bushmeat market in Malabo. Key Words: Bioko Island, body measurements, conservation, monkeys, morphology, taxonomy Introduction gordonorum), and surprisingly few such data exist even for some of the more widespread species (for example, Allen’s Comparing external body measurements for adult indi- swamp monkey Allenopithecus nigroviridis, northern tala- viduals from different sites has long been used as a tool for poin monkey Miopithecus ogouensis, and grivet Chlorocebus describing populations, subspecies, and species of animals aethiops). -

Assessment of Population Density and Structure of Primates in Pandam Wildlife Park, Plateau State, Nigeria
Sustainability, Agri, Food and Environmental Research, (ISSN: 0719-3726) , 6(2), 2018: 18-35 18 http://dx.doi.org/10.7770/safer-V6N2-art1503 ASSESSMENT OF POPULATION DENSITY AND STRUCTURE OF PRIMATES IN PANDAM WILDLIFE PARK, PLATEAU STATE, NIGERIA DENSIDAD POBLACIONAL Y ESTRUCTURA DE PRIMATES DIURNOS EN EL PARQUE DE VIDA SILVESTRE EN PANDAM, ESTADO DE PLATEAU, NIGERIA. Gabriel Ortyom Yager*, James Oshita Bukie and Avalumun Emmanuel Kaa, Department of Wildlife and Range Management, University of Agriculture, Makurdi, Benue State, Nigeria. *Correspondent author email & Phone: [email protected]; 08150609846 ABSTRACT A survey of diurnal primate species in Pandam Wildlife Park, Nigeria was conducted to determine its population density and structure. Eight transect lines (2.0km length, 0.02km width) at interval of 1.0km were located as representative samples in the Park within the Three-range stratum (riparian forest, savannah woodland and, swampy area) based on proportional to size of the strata in providing information on the primates’ species present in the Park which include Cercopithecus mona, Erythrocebus patas, Papio anubis and, Chlorocebus tantalus. Direct method of animal sighting was employed. Data was collected and analyzed using descriptive statistics, ANOVA and diversity indices. The results showed that savannah woodland strata had more number of individual species encountered (132) and the lowest was the swampy area. Also the savannah woodland had the highest species diversity and richness while the riparian forest strata had the highest number of species evenness. More so, Cercopithecus tantalus was widespread throughout the Park among other primates and Cercopithecus mona is most likely to decline even more rapidly than others since they inhabit the very tall trees. -

The Taxonomy of Primates in the Laboratory Context
P0800261_01 7/14/05 8:00 AM Page 3 C HAPTER 1 The Taxonomy of Primates T HE T in the Laboratory Context AXONOMY OF P Colin Groves RIMATES School of Archaeology and Anthropology, Australian National University, Canberra, ACT 0200, Australia 3 What are species? D Taxonomy: EFINITION OF THE The biological Organizing nature species concept Taxonomy means classifying organisms. It is nowadays commonly used as a synonym for systematics, though Disagreement as to what precisely constitutes a species P strictly speaking systematics is a much broader sphere is to be expected, given that the concept serves so many RIMATE of interest – interrelationships, and biodiversity. At the functions (Vane-Wright, 1992). We may be interested basis of taxonomy lies that much-debated concept, the in classification as such, or in the evolutionary implica- species. tions of species; in the theory of species, or in simply M ODEL Because there is so much misunderstanding about how to recognize them; or in their reproductive, phys- what a species is, it is necessary to give some space to iological, or husbandry status. discussion of the concept. The importance of what we Most non-specialists probably have some vague mean by the word “species” goes way beyond taxonomy idea that species are defined by not interbreeding with as such: it affects such diverse fields as genetics, biogeog- each other; usually, that hybrids between different species raphy, population biology, ecology, ethology, and bio- are sterile, or that they are incapable of hybridizing at diversity; in an era in which threats to the natural all. Such an impression ultimately derives from the def- world and its biodiversity are accelerating, it affects inition by Mayr (1940), whereby species are “groups of conservation strategies (Rojas, 1992). -

Conservation News
Conservation news New species of monkey discovered in www.redlist.org/info/categories_criteria2001.html), the Tanzania: the Critically Endangered major criteria being its extremely limited distribution, highland mangabey Lophocebus kipunji fragmentation into two populations, and the likelihood that its abundance is low. In Ndundulu Forest in the A new species of monkey, the first to be discovered in Udzungwas the major concern is that there are likely to Africa since 1984, has been unexpectedly found in two be very few Highland mangabeys remaining, perhaps separate montane forest areas in Tanzania: Ndundulu fewer than 500 animals. Our preliminary surveys found Forest of the Udzungwa Mountains and Rungwe- only three groups, all within a 3 km2 area; previous Livingstone in the Southern Highlands, 350 km from surveys in this forest and discussions with local villagers Ndundulu. The monkey is a mangabey and has been indicated that the mangabey is absent from much of named the Highland mangabey Lophocebus kipunji. The Ndundulu Forest. The forest is in good condition, common name reflects the fact that it is found in forests although its status of Forest Reserve, as it is now classi- at elevations above 1,300 m and as high as 2,450 m, fied, does not provide the degree of monitoring and - where temperatures can drop to 3°C. The species protection that will fully assure survival of this small name, kipunji (pronounced kip-oon-jee), is the name that population of the mangabey. We are continuing efforts local people in the Southern Highlands have for the ‘shy to encourage the Tanzanian government to incorporate monkey’ they reported seeing from time to time in the Ndundulu Forest Reserve into adjacent Udzungwa forest. -

2016 Activities Report Wapca in Action Creating Viable Long-Term Solutions
Page 0 of 28 WEST WEST AFRICAN AFRICAN PRIMATE PRIMATE CONSERVATION CONSERVATION ACTION ACTION 2015 Annual Report Annual Report 2016 Annual Report West African Primate Conservation Action Annual Report 2016 Page 1 of 28 WEST AFRICAN PRIMATE CONSERVATION ACTION 2016 ANNUAL REPORT Andrea Dempsey. BA. MSc. Country Coordinator West African Primate Conservation Action P.O. Box GP1319 Accra, Ghana Email: [email protected] Website: www.wapca.org West African Primate Conservation Action Annual Report 2016 Page 2 of 28 Update from the WAPCA-Ghana Country Coordinator WAPCA members, sponsors, partners and friends have continued to generously support us over the past year allowing us to continue the work here in Ghana at both the Critically Endangered Primate Breeding Centre, Kumasi Zoo and also in the field working with communities to protect the forests and the wild primates that inhabit them, for which we are hugely grateful. We’d like to particularly welcome our newest WAPCA member Africa Alive. WAPCA Ghana Board also grew with five new Board members joining – Dr Selorm Tettey, Edward Wiafe, Kwame Tutu, Micheal Abedi-Lartey and Noah Gbexede. The Board also elected David Tettey as our new Chairman. David has been Acting Chairman since the sad passing of our late Chairman – we look forward to him guiding us over the coming years. 2016 saw many visitors – David Morgan from Colchester Zoo, Heather MacIntosh, Hannah Joy and Miranda Crosby from ZSL London Zoo, Tobias Kremer from Heidelberg Zoo, Sabrina Linn from Frankfurt Zoo and Nicky Plaskitt from Paradise Park – I wish to thank them for spending time with our team here and sharing their expertise and knowledge, as well as the useful gifts which they donated including enrichment items donated from Zoo Wizards. -

The Behavioral Ecology of the Tibetan Macaque
Fascinating Life Sciences Jin-Hua Li · Lixing Sun Peter M. Kappeler Editors The Behavioral Ecology of the Tibetan Macaque Fascinating Life Sciences This interdisciplinary series brings together the most essential and captivating topics in the life sciences. They range from the plant sciences to zoology, from the microbiome to macrobiome, and from basic biology to biotechnology. The series not only highlights fascinating research; it also discusses major challenges associ- ated with the life sciences and related disciplines and outlines future research directions. Individual volumes provide in-depth information, are richly illustrated with photographs, illustrations, and maps, and feature suggestions for further reading or glossaries where appropriate. Interested researchers in all areas of the life sciences, as well as biology enthu- siasts, will find the series’ interdisciplinary focus and highly readable volumes especially appealing. More information about this series at http://www.springer.com/series/15408 Jin-Hua Li • Lixing Sun • Peter M. Kappeler Editors The Behavioral Ecology of the Tibetan Macaque Editors Jin-Hua Li Lixing Sun School of Resources Department of Biological Sciences, Primate and Environmental Engineering Behavior and Ecology Program Anhui University Central Washington University Hefei, Anhui, China Ellensburg, WA, USA International Collaborative Research Center for Huangshan Biodiversity and Tibetan Macaque Behavioral Ecology Anhui, China School of Life Sciences Hefei Normal University Hefei, Anhui, China Peter M. Kappeler Behavioral Ecology and Sociobiology Unit, German Primate Center Leibniz Institute for Primate Research Göttingen, Germany Department of Anthropology/Sociobiology University of Göttingen Göttingen, Germany ISSN 2509-6745 ISSN 2509-6753 (electronic) Fascinating Life Sciences ISBN 978-3-030-27919-6 ISBN 978-3-030-27920-2 (eBook) https://doi.org/10.1007/978-3-030-27920-2 This book is an open access publication. -

Primate Connections
Primate Connections !"#! Foreword by Dr. Simon Bearder Cover Photo by Noel Rowe THANK YOU! ear Friends, To complete the conservation circle, this DThank you for your support! By purchasing this calendar, you calendar also builds capacity in the places have become part of the Primate Connection – a network of grass where primates need it most. Part of the roots primate conservation organizations working collectively to proceeds from the sale of this calendar are raise awareness about the plight of primates, while generating sent to Oxford Brookes University’s Pri innovative programs that are helping to protect primates throughout mate HabitatCountry Student Scholarship. the world. © Sam Trull This scholarship grants students who come ! "#$%!&'&()*'+,!-'.!/00!10#,.*02!%0*0!3#/!#/402!,'!/%#*0!35,%! from countries where primates live a chance us a synopsis of the conservation issues they face and the creative to receive a Master’s degree in Primate Conservation Biology. ways they go about tackling them. Their renditions, along with A heartfelt thank you is given to those individuals from each photos of some of the rarest and most beautiful primates on the organization that made this project possible: Andrea Donaldson, planet, were then compiled to make this calendar. Helen Thirlway, Shirley McGreal, Steve Coan, Marc Myers, Noel As you read through the pages, you’ll become familiar with Rowe, Lis Key, Aura BeckhoferFialho, Sam Trull, Liz Tyson, Sam some of the most dedicated primate conservation organizations in Shanee, Joy Lliff, Aoife Healy, Pedro MendezCarvajal, Dominique the world. The spectrum of mitigation strategies they employ is both Aubin, Steve Blumkin, Brook Aldrich, Petra Osterberg, Sarah Rus inspiring and vital. -
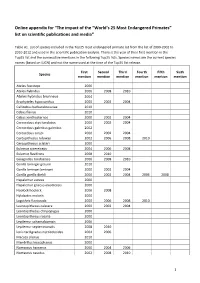
Online Appendix for “The Impact of the “World's 25 Most Endangered
Online appendix for “The impact of the “World’s 25 Most Endangered Primates” list on scientific publications and media” Table A1. List of species included in the Top25 most endangered primate list from the list of 2000-2002 to 2010-2012 and used in the scientific publication analysis. There is the year of their first mention in the Top25 list and the consecutive mentions in the following Top25 lists. Species names are the current species names (based on IUCN) and not the name used at the time of the Top25 list release. First Second Third Fourth Fifth Sixth Species mention mention mention mention mention mention Ateles fusciceps 2006 Ateles hybridus 2006 2008 2010 Ateles hybridus brunneus 2004 Brachyteles hypoxanthus 2000 2002 2004 Callicebus barbarabrownae 2010 Cebus flavius 2010 Cebus xanthosternos 2000 2002 2004 Cercocebus atys lunulatus 2000 2002 2004 Cercocebus galeritus galeritus 2002 Cercocebus sanjei 2000 2002 2004 Cercopithecus roloway 2002 2006 2008 2010 Cercopithecus sclateri 2000 Eulemur cinereiceps 2004 2006 2008 Eulemur flavifrons 2008 2010 Galagoides rondoensis 2006 2008 2010 Gorilla beringei graueri 2010 Gorilla beringei beringei 2000 2002 2004 Gorilla gorilla diehli 2000 2002 2004 2006 2008 Hapalemur aureus 2000 Hapalemur griseus alaotrensis 2000 Hoolock hoolock 2006 2008 Hylobates moloch 2000 Lagothrix flavicauda 2000 2006 2008 2010 Leontopithecus caissara 2000 2002 2004 Leontopithecus chrysopygus 2000 Leontopithecus rosalia 2000 Lepilemur sahamalazensis 2006 Lepilemur septentrionalis 2008 2010 Loris tardigradus nycticeboides -

REVIEW ARTICLE Agroecosystems and Primate Conservation in the Tropics: a Review
American Journal of Primatology 74:696–711 (2012) REVIEW ARTICLE Agroecosystems and Primate Conservation in The Tropics: A Review ∗ ALEJANDRO ESTRADA1 , BECKY E. RABOY2,3, AND LEONARDO C. OLIVEIRA3-6 1Estaci´on de Biolog´ıa Tropical Los Tuxtlas Instituto de Biolog´ıa, Universidad Nacional Aut´onoma de M´exico, Mexico City, Mexico 2Conservation Ecology Center, Smithsonian Conservation Biology Institute, National Zoological Park, Washington, DC 3Instituto de Estudos S´ocioambientais do Sul da Bahia (IESB), Ilh´eus-BA, Brazil 4Programa de P´os-Gradua¸c˜ao em Ecologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 5Programa de P´os-Gradua¸c˜ao em Ecologia e Conserva¸c˜ao da Biodiversidade, Universidade Estadual de Santa Cruz, Ilh´eus-BA, Brazil 6Bicho do Mato Instituto de Pesquisa, Belo Horizonte-MG, Brazil Agroecosystems cover more than one quarter of the global land area (ca. 50 million km2) as highly simplified (e.g. pasturelands) or more complex systems (e.g. polycultures and agroforestry systems) with the capacity to support higher biodiversity. Increasingly more information has been published about primates in agroecosystems but a general synthesis of the diversity of agroecosystems that primates use or which primate taxa are able to persist in these anthropogenic components of the landscapes is still lacking. Because of the continued extensive transformation of primate habitat into human-modified landscapes, it is important to explore the extent to which agroecosystems are used by primates. In this article, we reviewed published information on the use of agroecosystems by primates in habitat countries and also discuss the potential costs and benefits to human and nonhuman primates of primate use of agroecosystems. -

High-Ranking Geladas Protect and Comfort Others After Conflicts
www.nature.com/scientificreports OPEN High-Ranking Geladas Protect and Comfort Others After Conficts Elisabetta Palagi1, Alessia Leone1, Elisa Demuru1 & Pier Francesco Ferrari2 Post-confict afliation is a mechanism favored by natural selection to manage conficts in animal Received: 2 January 2018 groups thus avoiding group disruption. Triadic afliation towards the victim can reduce the likelihood Accepted: 30 August 2018 of redirection (benefts to third-parties) and protect and provide comfort to the victim by reducing its Published: xx xx xxxx post-confict anxiety (benefts to victims). Here, we test specifc hypotheses on the potential functions of triadic afliation in Theropithecus gelada, a primate species living in complex multi-level societies. Our results show that higher-ranking geladas provided more spontaneous triadic afliation than lower- ranking subjects and that these contacts signifcantly reduced the likelihood of further aggression on the victim. Spontaneous triadic afliation signifcantly reduced the victim’s anxiety (measured by scratching), although it was not biased towards kin or friends. In conclusion, triadic afliation in geladas seems to be a strategy available to high-ranking subjects to reduce the social tension generated by a confict. Although this interpretation is the most parsimonious one, it cannot be totally excluded that third parties could also be afected by the negative emotional state of the victim thus increasing a third party’s motivation to provide comfort. Therefore, the debate on the linkage between third-party afliation and emotional contagion in monkeys remains to be resolved. Conficts in social animals can have various immediate and long-term outcomes. Immediately following a con- fict, opponents may show a wide range of responses, from tolerance and avoidance of open confict, to aggres- sion1. -

Population Composition and Density of Mona Monkey in Lekki Concervation Centre, Lekki, Lagos, Nigeria
Olaleru et al., 2020 Journal of Research in Forestry, Wildlife & Environment Vol. 12(3) September, 2020 E-mail: [email protected]; [email protected] http://www.ajol.info/index.php/jrfwe This work is licensed under a jfewr ©2020 - jfewr Publications 259 Creative Commons Attribution 4.0 License ISBN: 2141 – 1778 Olaleru et al., 2020 POPULATION COMPOSITION AND DENSITY OF MONA MONKEY IN LEKKI CONCERVATION CENTRE, LEKKI, LAGOS, NIGERIA *1, 2Olaleru, F., 1Omotosho, O. O. and 1, 2Omoregie, Q. O. 1Department of Zoology, Faculty of Science, University of Lagos, Lagos State, Nigeria. 2Centre for Biodiversity Conservation and Ecosystem Management, University of Lagos, Lagos State, Nigeria. * Corresponding Author: [email protected]; + 234 807 780 0748 ABSTRACT The mona monkey (Cercopitecus mona) is the only non-human primate in Lekki Conservation Centre (LCC), a 78 hectares Strict Nature Reserve located in a peri-urban part of Lagos, Nigeria. This study aimed to of population composition and density of mona monkeys in LCC. Total count method using woods walk ways and perimeter road as line transects was used for the enumeration. The censuses were conducted for 27 days in October, November, and December, 2018. Counts were carried out between 06:30 and 10:30 hours for 22 days, and between 16:30 and 19:00 hours for five days. Monkeys were enumerated by counting all sighted individuals on both sides of the walk ways, and other study points. Data was subjected to analysis of variance to compare the monthly means, and statistically significant means (P < 0.05) were separated using Tukey post hoc test.