Rlab + Rlabplus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fortran Resources 1
Fortran Resources 1 Ian D Chivers Jane Sleightholme May 7, 2021 1The original basis for this document was Mike Metcalf’s Fortran Information File. The next input came from people on comp-fortran-90. Details of how to subscribe or browse this list can be found in this document. If you have any corrections, additions, suggestions etc to make please contact us and we will endeavor to include your comments in later versions. Thanks to all the people who have contributed. Revision history The most recent version can be found at https://www.fortranplus.co.uk/fortran-information/ and the files section of the comp-fortran-90 list. https://www.jiscmail.ac.uk/cgi-bin/webadmin?A0=comp-fortran-90 • May 2021. Major update to the Intel entry. Also changes to the editors and IDE section, the graphics section, and the parallel programming section. • October 2020. Added an entry for Nvidia to the compiler section. Nvidia has integrated the PGI compiler suite into their NVIDIA HPC SDK product. Nvidia are also contributing to the LLVM Flang project. Updated the ’Additional Compiler Information’ entry in the compiler section. The Polyhedron benchmarks discuss automatic parallelisation. The fortranplus entry covers the diagnostic capability of the Cray, gfortran, Intel, Nag, Oracle and Nvidia compilers. Updated one entry and removed three others from the software tools section. Added ’Fortran Discourse’ to the e-lists section. We have also made changes to the Latex style sheet. • September 2020. Added a computer arithmetic and IEEE formats section. • June 2020. Updated the compiler entry with details of standard conformance. -

With SCL and C/C++ 3 Graphics for Guis and Data Plotting 4 Implementing Computational Models with C and Graphics José M
Presentation Agenda Using Cross-Platform Graphic Packages for 1 General Goals GUIs and Data Plotting 2 Concepts and Background Overview with SCL and C/C++ 3 Graphics for GUIs and data plotting 4 Implementing Computational Models with C and graphics José M. Garrido C. 5 Demonstration of C programs using graphic libraries on Linux Department of Computer Science College of Science and Mathematics cs.kennesaw.edu/~jgarrido/comp_models Kennesaw State University November 7, 2014 José M. Garrido C. Graphic Packages for GUIs and Data Plotting José M. Garrido C. Graphic Packages for GUIs and Data Plotting A Computational Model Abstraction and Decomposition An software implementation of the solution to a (scientific) Abstraction is recognized as a fundamental and essential complex problem principle in problem solving and software development. It usually requires a mathematical model or mathematical Abstraction and decomposition are extremely important representation that has been formulated for the problem. in dealing with large and complex systems. The software implementation often requires Abstraction is the activity of hiding the details and exposing high-performance computing (HPC) to run. only the essential features of a particular system. In modeling, one of the critical tasks is representing the various aspects of a system at different levels of abstraction. A good abstraction captures the essential elements of a system, and purposely leaving out the rest. José M. Garrido C. Graphic Packages for GUIs and Data Plotting José M. Garrido C. Graphic Packages for GUIs and Data Plotting Developing a Computational Model Levels of Abstraction in the Process The process of developing a computational model can be divided in three levels: 1 Prototyping, using the Matlab programming language 2 Performance implementation, using: C or Fortran programming languages. -
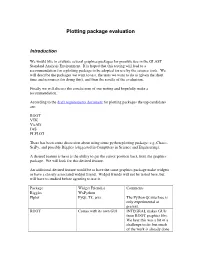
Plotting Package Evaluation
Plotting package evaluation Introduction We would like to evaluate several graphics packages for possible use in the GLAST Standard Analysis Environment. It is hoped that this testing will lead to a recommendation for a plotting package to be adopted for use by the science tools. We will describe the packages we want to test, the tests we want to do to (given the short time and resources for doing this), and then the results of the evaluation. Finally we will discuss the conclusions of our testing and hopefully make a recommendation. According to the draft requirements document for plotting packages the top candidates are: ROOT VTK VisAD JAS PLPLOT There has been some discussion about using some python plotting package: e.g.,Chaco, SciPy, and possibly Biggles (suggested in Computers in Science and Engineering). A desired feature is have is the ability to get the cursor position back from the graphics package. We will look for this desired feature. An additional desired feature would be to have the same graphics package make widgets or have a closely associated widget friend. Widget friends will not be tested here, but will have to studied before agreeing to use it. Package Widget Friend(s) Comments Biggles WxPython Plplot PyQt, Tk, java The Python Qt interface is only experimental at present. ROOT Comes with its own GUI INTEGRAL makes GUIs from ROOT graphics libs. We hear this was a bit of a challenge to do, but much of the work is already done for us. for us. Tests: The testing is to be carried out separately in the Windows and Linux environments. -

Volume 30 Number 1 March 2009
ADA Volume 30 USER Number 1 March 2009 JOURNAL Contents Page Editorial Policy for Ada User Journal 2 Editorial 3 News 5 Conference Calendar 30 Forthcoming Events 37 Articles J. Barnes “Thirty Years of the Ada User Journal” 43 J. W. Moore, J. Benito “Progress Report: ISO/IEC 24772, Programming Language Vulnerabilities” 46 Articles from the Industrial Track of Ada-Europe 2008 B. J. Moore “Distributed Status Monitoring and Control Using Remote Buffers and Ada 2005” 49 Ada Gems 61 Ada-Europe Associate Members (National Ada Organizations) 64 Ada-Europe 2008 Sponsors Inside Back Cover Ada User Journal Volume 30, Number 1, March 2009 2 Editorial Policy for Ada User Journal Publication Original Papers Commentaries Ada User Journal — The Journal for Manuscripts should be submitted in We publish commentaries on Ada and the international Ada Community — is accordance with the submission software engineering topics. These published by Ada-Europe. It appears guidelines (below). may represent the views either of four times a year, on the last days of individuals or of organisations. Such March, June, September and All original technical contributions are articles can be of any length – December. Copy date is the last day of submitted to refereeing by at least two inclusion is at the discretion of the the month of publication. people. Names of referees will be kept Editor. confidential, but their comments will Opinions expressed within the Ada Aims be relayed to the authors at the discretion of the Editor. User Journal do not necessarily Ada User Journal aims to inform represent the views of the Editor, Ada- readers of developments in the Ada The first named author will receive a Europe or its directors. -

CC Data Visualization Library
CC Data Visualization Library: Visualizing large amounts of scientific data through interactive graph widgets on ordinary workstations Master of Science Thesis in the programmes Interaction Design & Algorithms, Languages and Logic MAX OCKLIND ERIK WIKLUND Chalmers University of Technology Department of Computer Science and Engineering Göteborg, Sweden, May 2012 The Authors grant to Chalmers University of Technology the non-exclusive right to publish the Work electronically and in a non-commercial purpose make it accessible on the Internet. The Authors warrant that they are the authors to the Work, and warrant that the Work does not contain text, pictures or other material that violates copyright law. The Authors shall, when transferring the rights of the Work to a third party (for example a publisher or a company), acknowledge the third party about this agreement. If the Authors have signed a copyright agreement with a third party regarding the Work, the Authors warrant hereby that they have obtained any necessary permission from this third party to let Chalmers University of Technology store the Work electronically and make it accessible on the Internet. CC Data Visualization Library: Visualizing large amounts of scientific data through interactive graph widgets on ordinary workstations MAX OCKLIND ERIK WIKLUND © MAX OCKLIND, May 2012. © ERIK WIKLUND, May 2012. Examiner: Olof Torgersson Chalmers University of Technology Department of Computer Science and Engineering SE-412 96 Göteborg Sweden Telephone + 46 (0)31-772 1000 Cover: The GUI of the CCDVL library during a test run, showing a scatter plot graph of sample data with a lasso selection overlay. Sections 4.2.3 FRONTEND AND GUI ANALYSIS AND DESIGN and 4.3.3 FRONTEND AND GUI IMPLEMENTATION as well as APPENDIX B - MANUAL AND USER GUIDE contains detailed descriptions of the GUI and frontend. -

Getting Started with Matlab (In Computer Science at UBC)
Getting Started with Matlab (in Computer Science at UBC) Ian Mitchell Department of Computer Science The University of British Columbia Outline • Why Matlab? – Why not C / C++ / Java / Fortran? – Why not Perl / Python? – Why not Mathematica / Maple? • A Brief Taste of Matlab – where to find it – how to run it – interactive Matlab – m-files & debugging • Sources of Additional Information January 2013 Ian Mitchell (UBC Computer Science) 2 Choosing Programming Languages • Compare / contrast compiled languages C++ and Java C++ Java • Fast: close to hardware • Easy to use: references only, • Flexible: interfaces to almost garbage collection any other language • Popular: commonly used, widely • Flexible: pointers, references, available explicit memory allocation • Portable: common byte code • Flexible: everybody provides C / • Developed with a clear vision: C++ libraries Standard libraries for security, • Popular: commonly used, threading, distributed systems available everywhere • Slower: interpreted or JIT for • Prone to bugs: complex syntax, byte code memory leaks January 2013 Ian Mitchell (UBC Computer Science) 3 The Right Tool for the Job • C / C++ / Fortran: – Statically typed and compiled languages – Well developed algorithm, known platform, execution time is key • Java: – Simpler, partially compiled language – Unknown platform, less experienced programmer, development time is important, broad standard library • Perl / Python: – Interpreted “dynamic” languages: no typing, no compilation(?) – Unknown platform, development time is key, -

Econometrics with Octave
Econometrics with Octave Dirk Eddelb¨uttel∗ Bank of Montreal, Toronto, Canada. [email protected] November 1999 Summary GNU Octave is an open-source implementation of a (mostly Matlab compatible) high-level language for numerical computations. This review briefly introduces Octave, discusses applications of Octave in an econometric context, and illustrates how to extend Octave with user-supplied C++ code. Several examples are provided. 1 Introduction Econometricians sweat linear algebra. Be it for linear or non-linear problems of estimation or infer- ence, matrix algebra is a natural way of expressing these problems on paper. However, when it comes to writing computer programs to either implement tried and tested econometric procedures, or to research and prototype new routines, programming languages such as C or Fortran are more of a bur- den than an aid. Having to enhance the language by supplying functions for even the most primitive operations adds extra programming effort, introduces new points of failure, and moves the level of abstraction further away from the elegant mathematical expressions. As Eddelb¨uttel(1996) argues, object-oriented programming provides `a step up' from Fortran or C by enabling the programmer to seamlessly add new data types such as matrices, along with operations on these new data types, to the language. But with Moore's Law still being validated by ever and ever faster processors, and, hence, ever increasing computational power, the prime reason for using compiled code, i.e. speed, becomes less relevant. Hence the growing popularity of interpreted programming languages, both, in general, as witnessed by the surge in popularity of the general-purpose programming languages Perl and Python and, in particular, for numerical applications with strong emphasis on matrix calculus where languages such as Gauss, Matlab, Ox, R and Splus, which were reviewed by Cribari-Neto and Jensen (1997), Cribari-Neto (1997) and Cribari-Neto and Zarkos (1999), have become popular. -

The Graphics Cookbook SC/15.2 —Abstract Ii
SC/15.2 Starlink Project STARLINK Cookbook 15.2 A. Allan, D. Terrett 22nd August 2000 The Graphics Cookbook SC/15.2 —Abstract ii Abstract This cookbook is a collection of introductory material covering a wide range of topics dealing with data display, format conversion and presentation. Along with this material are pointers to more advanced documents dealing with the various packages, and hints and tips about how to deal with commonly occurring graphics problems. iii SC/15.2—Contents Contents 1 Introduction 3 2 Call for contributions 3 3 Subroutine Libraries 3 3.1 The PGPLOT library . 3 3.1.1 Encapsulated Postscript and PGPLOT . 5 3.1.2 PGPLOT Environment Variables . 5 3.1.3 PGPLOT Postscript Environment Variables . 5 3.1.4 Special characters inside PGPLOT text strings . 7 3.2 The BUTTON library . 7 3.3 The pgperl package . 11 3.3.1 Argument mapping – simple numbers and arrays . 12 3.3.2 Argument mapping – images and 2D arrays . 13 3.3.3 Argument mapping – function names . 14 3.3.4 Argument mapping – general handling of binary data . 14 3.4 Python PGPLOT . 14 3.5 GLISH PGPLOT . 15 3.6 ptcl Tk/Tcl and PGPLOT . 15 3.7 Starlink/Native PGPLOT . 15 3.8 Graphical Kernel System (GKS) . 16 3.8.1 Enquiring about the display . 16 3.8.2 Compiling and Linking GKS programs . 17 3.9 Simple Graphics System (SGS) . 17 3.10 PLplot Library . 17 3.10.1 PLplot and 3D Surface Plots . 18 3.11 The libjpeg Library . 19 3.12 The giflib Library . -

Documentation of the Plplot Plotting Software I
Documentation of the PLplot plotting software i Documentation of the PLplot plotting software Documentation of the PLplot plotting software ii Copyright © 1994 Maurice J. LeBrun, Geoffrey Furnish Copyright © 2000-2005 Rafael Laboissière Copyright © 2000-2016 Alan W. Irwin Copyright © 2001-2003 Joao Cardoso Copyright © 2004 Andrew Roach Copyright © 2004-2013 Andrew Ross Copyright © 2004-2016 Arjen Markus Copyright © 2005 Thomas J. Duck Copyright © 2005-2010 Hazen Babcock Copyright © 2008 Werner Smekal Copyright © 2008-2016 Jerry Bauck Copyright © 2009-2014 Hezekiah M. Carty Copyright © 2014-2015 Phil Rosenberg Copyright © 2015 Jim Dishaw Redistribution and use in source (XML DocBook) and “compiled” forms (HTML, PDF, PostScript, DVI, TeXinfo and so forth) with or without modification, are permitted provided that the following conditions are met: 1. Redistributions of source code (XML DocBook) must retain the above copyright notice, this list of conditions and the following disclaimer as the first lines of this file unmodified. 2. Redistributions in compiled form (transformed to other DTDs, converted to HTML, PDF, PostScript, and other formats) must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. Important: THIS DOCUMENTATION IS PROVIDED BY THE PLPLOT PROJECT “AS IS” AND ANY EXPRESS OR IM- PLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE PLPLOT PROJECT BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS DOCUMENTATION, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. -

Basic Seismic Utilities User's Guide
Basic Seismic Utilities User's Guide Dr. P. Michaels, PE 30 April 2002 <[email protected]> Release 1.0 Version 1 Software for Engineering Geophysics Center for Geophysical Investigation of the Shallow Subsurface Boise State University Boise, Idaho 83725 2 Acknowledgements I have built on the work of many others in the development of this package. I would like to thank Enders A. Robinson and the Holden-Day Inc., Liquidation Trust (1259 S.W. 14th Street, Boca Raton, FL 33486, Phone: 561.750-9229 Fax: 561.394.6809) for license to include and distribute under the GNU license subroutines found in Dr. Robinson’s 1967 book [14] , Multichannel time series analysis with digital computer programs. This book is currently out of print, but contains a wealth of algorithms, several of which I have found useful and included in the BSU Fortran77 subroutine library (sublib4.a). This has saved me considerable time. In other cases, subroutines taken from the book Numerical Recipes [12] had to be replaced (the publisher did not give permission to distribute). While this is an excellent book, and very instructional for those interested in the theory of the algorithms, future authors of software should know that the algorithms given in that book are NOT GNU. Replacement software was found in the GNU Scientific Library (GSL), and in the CMLIB. The original plotting software was from PGPLOT (open source, but distribution restricted). These codes have since been replaced with GNU compliant PLPLOT routines. PLPLOT credits include Dr. Maurice LeBrun <[email protected]> and Geoffrey Furnish <[email protected]>. -

Development of a GUI-Wrapper MPX for Reactor Physics Design and Verification
Transactions of the Korean Nuclear Society Autumn Meeting Gyeongju, Korea, October 25-26, 2012 Development of a GUI-Wrapper MPX for Reactor Physics Design and Verification In Seob Hong * and Sweng Woong Woo Korea Institute of Nuclear Safety 62 Gwahak-ro, Yuseong-gu, Daejeon, 305-338, Republic of Korea *Corresponding author: [email protected] 1. Introduction are inter-compiled within script-based languages such as Perl, Python, Tcl, Java, Ruby, etc. More recently but for a few decades there has been a In the present work, the PLplot library has been lot of interest in the graphical user interface (GUI) for chosen to develop the MPX wrapper program through engineering design, analysis and verification purposes. a wide range of literature survey and toy experiments. For nuclear engineering discipline, the situation would not be much different. 3. Development of MPX; a GUI-Wrapper The advantages of developing a GUI-based computer program might be sought from two aspects; a) efficient 2.1 Summary of PLplot Library verification of geometrical models and numerical results from designers’ point of view, and b) quick- PLplot is a cross-platform software package for hand and independent verification of designers’ works creating scientific plots written basically in C language. from regulators’ point view. As the other GUI library package, the PLplot core The objective of this paper is to present a GUI- library can be used to create standard x-y plots, semi- wrapper program named as MPX, which is intended to log plots, log-log plots, contour plots, 3D surface plots, be efficient but not heavy. -

Documentation of the Plplot Plotting Software I
Documentation of the PLplot plotting software i Documentation of the PLplot plotting software Documentation of the PLplot plotting software ii Copyright © 1994 Maurice J. LeBrun, Geoffrey Furnish Copyright © 2000-2005 Rafael Laboissière Copyright © 2000-2016 Alan W. Irwin Copyright © 2001-2003 Joao Cardoso Copyright © 2004 Andrew Roach Copyright © 2004-2013 Andrew Ross Copyright © 2004-2016 Arjen Markus Copyright © 2005 Thomas J. Duck Copyright © 2005-2010 Hazen Babcock Copyright © 2008 Werner Smekal Copyright © 2008-2016 Jerry Bauck Copyright © 2009-2014 Hezekiah M. Carty Copyright © 2014-2015 Phil Rosenberg Copyright © 2015 Jim Dishaw Redistribution and use in source (XML DocBook) and “compiled” forms (HTML, PDF, PostScript, DVI, TeXinfo and so forth) with or without modification, are permitted provided that the following conditions are met: 1. Redistributions of source code (XML DocBook) must retain the above copyright notice, this list of conditions and the following disclaimer as the first lines of this file unmodified. 2. Redistributions in compiled form (transformed to other DTDs, converted to HTML, PDF, PostScript, and other formats) must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. Important: THIS DOCUMENTATION IS PROVIDED BY THE PLPLOT PROJECT “AS IS” AND ANY EXPRESS OR IM- PLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE PLPLOT PROJECT BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS DOCUMENTATION, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.