Population-Genetic Structure of a Philopatric, Colonially Nesting Seabird, the Short-Tailed Shearwater (Puffinus Tenuirostris)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
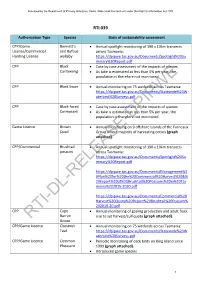
Rti-Dl-Release-Dpipwe
Assessed by the Department of Primary Industries, Parks, Water and Environment under the Right to Information Act 2009 RTI 039 Authorisation Type Species Basis of sustainability assessment CPP/Game Bennett’s • Annual spotlight monitoring of 190 x 10km transects Licence/Commercial and Rufous across Tasmania: Hunting Licence wallaby https://dpipwe.tas.gov.au/Documents/Spotlight%20Su mmary%20Report.pdf CPP Black • Case by case assessment of the impacts of species Currawong • As take is estimated at less than 5% per year, the population is therefore not monitored. CPP Black Swan • Annual monitoring on 75 wetlands across Tasmania: https://dpipwe.tas.gov.au/Documents/Statewide%20W aterbird%20Surveys.pdf CPP Black-faced • Case by case assessment of the impacts of species Cormorant • As take is estimated at less than 5% per year, the population is therefore not monitored. Game Licence Brown • Annual monitoring on 9 offshore islands of the Furneaux Quail Group where majority of harvesting occurs (graph attached). CPP/Commercial Brushtail • Annual spotlight monitoring of 190 x 10km transects possum across Tasmania: https://dpipwe.tas.gov.au/Documents/Spotlight%20Su mmary%20Report.pdf https://dpipwe.tas.gov.au/Documents/Management%2 0Plan%20for%20the%20Commercial%20Harvest%20&% 20Export%20of%20Brushtail%20Possums%20in%20Tas mania%202015-2020.pdf https://dpipwe.tas.gov.au/Documents/Commercial%20 Harvest%20Quota%20Report%20Brushtail%20Possum% 202019-20.pdf CPP Cape • Annual monitoring of gosling production and adult flock Barren size to set harvest/cull quota (graph attached). RTI-DL-RELEASE-DPIPWEGoose CPP/Game Licence Chestnut • Annual monitoring on 75 wetlands across Tasmania: Teal https://dpipwe.tas.gov.au/Documents/Statewide%20W aterbird%20Surveys.pdf CPP/Game Licence Common • Periodic monitoring of cock birds on King Island since Pheasant 1999 (graph attached). -

Great Australian Bight BP Oil Drilling Project
Submission to Senate Inquiry: Great Australian Bight BP Oil Drilling Project: Potential Impacts on Matters of National Environmental Significance within Modelled Oil Spill Impact Areas (Summer and Winter 2A Model Scenarios) Prepared by Dr David Ellis (BSc Hons PhD; Ecologist, Environmental Consultant and Founder at Stepping Stones Ecological Services) March 27, 2016 Table of Contents Table of Contents ..................................................................................................... 2 Executive Summary ................................................................................................ 4 Summer Oil Spill Scenario Key Findings ................................................................. 5 Winter Oil Spill Scenario Key Findings ................................................................... 7 Threatened Species Conservation Status Summary ........................................... 8 International Migratory Bird Agreements ............................................................. 8 Introduction ............................................................................................................ 11 Methods .................................................................................................................... 12 Protected Matters Search Tool Database Search and Criteria for Oil-Spill Model Selection ............................................................................................................. 12 Criteria for Inclusion/Exclusion of Threatened, Migratory and Marine -

Overview of Tasmania's Offshore Islands and Their Role in Nature
Papers and Proceedings of the Royal Society of Tasmania, Volume 154, 2020 83 OVERVIEW OF TASMANIA’S OFFSHORE ISLANDS AND THEIR ROLE IN NATURE CONSERVATION by Sally L. Bryant and Stephen Harris (with one text-figure, two tables, eight plates and two appendices) Bryant, S.L. & Harris, S. 2020 (9:xii): Overview of Tasmania’s offshore islands and their role in nature conservation.Papers and Proceedings of the Royal Society of Tasmania 154: 83–106. https://doi.org/10.26749/rstpp.154.83 ISSN: 0080–4703. Tasmanian Land Conservancy, PO Box 2112, Lower Sandy Bay, Tasmania 7005, Australia (SLB*); Department of Archaeology and Natural History, College of Asia and the Pacific, Australian National University, Canberra, ACT 2601 (SH). *Author for correspondence: Email: [email protected] Since the 1970s, knowledge of Tasmania’s offshore islands has expanded greatly due to an increase in systematic and regional surveys, the continuation of several long-term monitoring programs and the improved delivery of pest management and translocation programs. However, many islands remain data-poor especially for invertebrate fauna, and non-vascular flora, and information sources are dispersed across numerous platforms. While more than 90% of Tasmania’s offshore islands are statutory reserves, many are impacted by a range of disturbances, particularly invasive species with no decision-making framework in place to prioritise their management. This paper synthesises the significant contribution offshore islands make to Tasmania’s land-based natural assets and identifies gaps and deficiencies hampering their protection. A continuing focus on detailed gap-filling surveys aided by partnership restoration programs and collaborative national forums must be strengthened if we are to capitalise on the conservation benefits islands provide in the face of rapidly changing environmental conditions and pressure for future use. -

Tasmanian Aborigines in the Furneaux Group in the Nine Teenth Century—Population and Land
‘I hope you will be my frend’: Tasmanian Aborigines in the Furneaux Group in the nine teenth century—population and land tenure Irynej Skira Abstract This paper traces the history of settlement of the islands of the Furneaux Group in Bass Strait and the effects of government regulation on the long term settlements of Tasma nian Aboriginal people from the 1850s to the early 1900s. Throughout the nineteenth century the Aboriginal population grew slowly eventually constituting approximately 40 percent of the total population of the Furneaux Group. From the 1860s outsiders used the existing land title system to obtain possession of the islands. Aborigines tried to establish tenure through the same system, but could not compete because they lacked capital, and were disadvantaged by isolation in their communication with gov ernment. Further, the islands' use for grazing excluded Aborigines who rarely had large herds of stock and were generally not agriculturalists. The majority of Aborigines were forced to settle on Cape Barren Island, where they built homes on a reserve set aside for them. European expansion of settlement on Flinders Island finally completed the disen franchisement of Aboriginal people by making the Cape Barren Island enclave depend ent on the government. Introduction In December 1869 Thomas Mansell, an Aboriginal, applied to lease a small island. He petitioned the Surveyor-General, T hope you will be my Frend...I am one of old hands Her, and haf Cast and have large family and no hum'.1 Unfortunately, he could not raise £1 as down payment. Mansell's was one of the many attempts by Aboriginal people in the Furneaux Group to obtain valid leasehold or freehold and recognition of their long term occupation. -

The Vegetation and Flora of Three Hummock Island, Western Bass Strait
Papers and Proceedings of the Royal Society of Tasmania, Volume 131, 1997 37 THE VEGETATION AND FLORA OF THREE HUMMOCK ISLAND, WESTERN BASS STRAIT by Stephen Harris and Jayne Balmer (with one table, four text-figures, nine plates and an appendix) HARRIS, S. & BALMER, ]., 1997 (31 :viii): The vegetation and flora of Three Hummock Island, western Bass Strait. Pap. Proc. R. Soc. Tasm. 131: 37-56. ISSN 0800-4703. Parks and Wildlife Service, Department of Environment and Land Management, GPO Box 44A, Hobart, Tasmania, Australia 7001. A survey of the vegetation of Three Hummock Island Nature Reserve recorded 289 vascular higher plant species, 60 of which were introduced. Of the native flora, six are classified as rare or vulnerable. Clarke Island, at a similar latitude in Eastern Bass Strait, has a significantly richer flora, including an element of Mainland Australian/Bassian flora for which the island is the southernmost limit. In contrast, there are no species known from Three Hummock Island that do not occur on mainland Tasmania. The greater length of time during which the land bridge at the eastern end of Bass Strait was exposed is, therefore, reflected in the flora. Three Hummock Island was cut off for a much longer period with no land connections to the north, therefore has a more insular Tasmanian flora. Climatic differences may have exacerbated the contrast. Nine vegetation mapping communities are defined, the largest proportion of the island being covered by Myrtaceae-dominated scrub. The main changes in the vegetation since the time of European discovery have been the clearing of much of the relatively fertile calcareous sands for grazing and the consequent loss of most of the Eucalyptus viminalis forests, an increased fire frequency and the introduction of exotic plants. -

The Fisher Island Field Stat Ion-With an Account of Its Principal Fauna and Flora
PAPEHS AND .PROCEEDINGS tW 'THE ROYAL SOCIETY OF TASMANIA, V'OLl.lME 92 THE FISHER ISLAND FIELD STAT ION-WITH AN ACCOUNT OF ITS PRINCIPAL FAUNA AND FLORA By E. R. GUILER, D. L. SERVENTY AND J. H. WILLIS (WITH 2 PLATES AND 9 TEXT FTGURESj t GENERAL DESCRIPTION OF FISHER ISLAND AND ITS MUTTON~BIRD ROOKERIES * INTRODUCTION mately 0'75 acres. The shoreline measures about Fisher Island Oat. 40° 10' S., long. 148° 16' E.) 530 yards and the greatest length, from North is among the smallest of the archipelago of islands Point to South Point, is 150 yards. Its elevation is comprising the Furneaux Group in eastern Bass about 19 feet above spring high-water mark. Strait. It lies off the southern shoreline of the Like the other islands in the Furneaux Group, major island in the group, Flinders Island, in Fisher Island is part of the basement Devonian Adelaide Bay, a portion of Franklin Sound which granite which forms the hills and mountain ridges separates Flinders Island from Cape Barren Island in the archipelago. On Flinders Island the low (fig. 1). Its convenient location to Lady Barron lying plains are covered by Tertiary alluvium and (the main port of Flinders Island, about 220 yards sands, with calcareous deposits in restricted areas. distant), its proximity to important commercial Limited Siluro-Devonian quartzites and slates also mutton-birding islands and the presence of a small, occur, and in the northern part of Adelaide Bay, at easily handled nesting colony of mutton-birds Petrifaction Bay, are exposures of Tertiary vesicu (PufJinus tenuirostris (Temminck), made it the lar basalts. -

Alphabetical Table Of
TASMANIAN ACTS AND STATUTORY RULES TASMANIAN ACTS N – R AND STATUTORY RULES Nation Building and Jobs Plan Facilitation (Tasmania) Act 2009, No. 5 of 2009 (commenced 27 April 2009) Last consolidation: 31 December 2012 (includes changes under the Legislation Publication Act 1996 in force as at 31 December 2012) Amendments commenced in 2009 – 2016: Nation Building and Jobs Plan Facilitation (Tasmania) Act 2009, No. 5 of 2009 (commenced 31 December 2012) – the Act, except Pt. 1 (ss. 1-4) and s. 18 expired 31 December 2012 unless earlier by notice made by the Treasurer National Broadband Network (Tasmania) Act 2010, No. 48 of 2010 (commenced 21 December 2010) Last consolidation: 16 August 2017 (up to and including amendment by the Aboriginal Relics (Consequential Amendments) Act 2017 and changes under the Legislation Publication Act 1996 in force as at 16 August 2017) Amendments commenced in 2017: Building (Consequential Amendments) Act 2016, No. 12 of 2016 (commenced 1 January 2017) – amended s. 28(c) Aboriginal Relics (Consequential Amendments) Act 2017, No. 17 of 2017 (commenced 16 August 2017) – amended s. 28 National Energy Retail Law (Tasmania) Act 2012, No. 11 of 2012 (commenced 1 July 2012, see S.R. 2012, No. 49) Last consolidation: 1 June 2013 (up to and including amendment by the Electricity Reform (Implementation) Act 2013 and changes under the Legislation Publication Act 1996 in force as at 1 June 2013) Amendments commenced in 2012 – 2016: Electricity Reform (Implementation) Act 2013, No. 5 of 2013 (commenced 1 June 2013) – amended ss. 15 and 18; inserted 17A Regulations: National Energy Retail Law (Tasmania) Regulations 2012 (2012/51 amended by 2013/27) National Energy Retail Law (Tasmania) s. -

Reserve Listing
Reserve Summary Report NCA Reserves Number Area (ha) Total 823 2,901,596.09 CONSERVATION AREA 438 661,640.89 GAME RESERVE 12 20,389.57 HISTORIC SITE 30 16,051.47 NATIONAL PARK 19 1,515,793.29 NATURE RECREATION AREA 25 67,340.19 NATURE RESERVE 86 118,977.14 REGIONAL RESERVE 148 454,286.95 STATE RESERVE 65 47,116.57 Total General Plan Total 823 2,901,596.09 823 2,901,596.09 CONSERVATION AREA 438 661,640.89 438 661,640.89 GAME RESERVE 12 20,389.57 12 20,389.57 HISTORIC SITE 30 16,051.47 30 16,051.47 NATIONAL PARK 19 1,515,793.29 19 1,515,793.29 NATURE RECREATION A 25 67,340.19 25 67,340.19 NATURE RESERVE 86 118,977.14 86 118,977.14 REGIONAL RESERVE 148 454,286.95 148 454,286.95 STATE RESERVE 65 47,116.57 65 47,116.57 CONSERVATION AREA Earliest Previous mgmt Name Mgt_plan IUCN Area ha Location Notes Reservation Statutory Rules Reservation auth NCA Adamsfield Conservation Area Yes - WHA Statutory VI 5,376.25 Derwent Valley Historic mining area 27-Jun-1990 1990#78 subject to PWS True 25.12.96 SR 1996 Alma Tier Conservation Area No IV 287.31 Glamorgan-Spring 03-Jan-2001 Alma Tier PWS True Bay Forest Reserve Alpha Pinnacle Conservation Area GMP - Reserve Report V 275.50 Southern Midlands Dry sclerophyll forest 24-Jul-1996 subject to 25.12.96 PWS True SR 1996 #234 Anderson Islands Conservation Area No V 749.57 Flinders 06 Apr 2011 PWS True Ansons Bay Conservation Area GMP - Reserve Report VI 104.56 Break ODay Coastal 27-May-1983 yyyy#76 PWS True Ansons River Conservation Area No VI 93.77 Ansons Bay 17-Apr-2013 SR13 of 2013 PWS True Apex Point -

Appendix 7-2 Protected Matters Search Tool (PMST) Report for the Risk EMBA
Environment plan Appendix 7-2 Protected matters search tool (PMST) report for the Risk EMBA Stromlo-1 exploration drilling program Equinor Australia B.V. Level 15 123 St Georges Terrace PERTH WA 6000 Australia February 2019 www.equinor.com.au EPBC Act Protected Matters Report This report provides general guidance on matters of national environmental significance and other matters protected by the EPBC Act in the area you have selected. Information on the coverage of this report and qualifications on data supporting this report are contained in the caveat at the end of the report. Information is available about Environment Assessments and the EPBC Act including significance guidelines, forms and application process details. Report created: 13/09/18 14:02:20 Summary Details Matters of NES Other Matters Protected by the EPBC Act Extra Information Caveat Acknowledgements This map may contain data which are ©Commonwealth of Australia (Geoscience Australia), ©PSMA 2010 Coordinates Buffer: 1.0Km Summary Matters of National Environmental Significance This part of the report summarises the matters of national environmental significance that may occur in, or may relate to, the area you nominated. Further information is available in the detail part of the report, which can be accessed by scrolling or following the links below. If you are proposing to undertake an activity that may have a significant impact on one or more matters of national environmental significance then you should consider the Administrative Guidelines on Significance. World Heritage Properties: 11 National Heritage Places: 13 Wetlands of International Importance: 13 Great Barrier Reef Marine Park: None Commonwealth Marine Area: 2 Listed Threatened Ecological Communities: 14 Listed Threatened Species: 311 Listed Migratory Species: 97 Other Matters Protected by the EPBC Act This part of the report summarises other matters protected under the Act that may relate to the area you nominated. -

1982 United Nations List of National Parks and Protected
:2 I 1982 United Nations List of National Parks and Protected Areas List des Nations Unies des Parcs Nationaux et des Aires Protégées 1982 Prepared by the lUCN Commission on National Parks and Protected Areas Préparée par la Commission des Parcs Nationaux et des Aires Protégées de 1 'UICN UNEP/PNUE Published with the financial assistance of Unesco and in cooperation with UNEP as a contributor to the Global Environment Monitoring System Publiée avec l'aide financière de l 'Unesco et travaillant dans un but commun avec PNUE; une contribution au système mondiale de surveillance continuée 1 'environment INTERNATIONAL UNION FOR CONSERVATION OF NATURE AND NATURAL RESOURCES UNION INTERNATIONALE POUR LA CONSERVATION DE LA NATURE ET DE SES RESSOURCES 1196 Gland, Suisse 1982 ) © 1982 lUCN Unesco subvention 1981 - 1983 DG/7 . 6. 2/SUB . 14 (SSC ISBN 2-88032-409-2 Printed in Great Britain by Unwin Brothers Ltd., The Gresham Press, Old Woking, Surrey 1982 United Nations List of National Parks and Protected Areas List des Nations Unies des Parcs Nationaux et des Aires Protégées 1982 Prepared by the lUCN Commission on National Parks and Protected Areas Préparée par la Commission des Parcs Nationaux et des Aires Protégées de 1 'UICN Published with the financial assistance of Unesco and in cooperation with UNEP as a contributon to the Global Environment Monitoring System Publiée avec l'aide financière de 1 'Unesco et travaillant dans un but commun avec PNUE; une contribution au système mondiale de surveillance continuée 1 'environment INTERNATIONAL UNION -

List of References
43 LIST OF REFERENCES 1. Aboriginal Lands Act 1995. 2. Aboriginal Lands Amendment (Wybalenna) Act 1999. 3. Aboriginal Lands Amendment Bill 1999. 4. Aird MLC, Hon Michael, Hansard, Legislative Council, 30 November 1999. 5. Bacon MHA, Premier, Jim, Transcript of Briefing, 1 February 2000. 6. Bingham, Mr Richard, Chairman, Aboriginal Land and Cultural Issues Working Group, Transcript of Evidence, 1 February and 10 April 2000. 7. Britton, Mr Ross, Arthur-Pieman Coalition, Transcript of Evidence, 16 March 2000. 8. Brown, Mr Greg, Department of Premier and Cabinet, Transcript of Evidence, 1 February 2000. 9. Circular Head Council, Transcript of Evidence, 14 March 2000. 10. Clark, Mr John, Tasmanian Regional Aboriginal Council, Transcript of Evidence, 16 March 2000. 11. Commonwealth Document, www.ilc.gov.au 12. Cooper, Mrs Helen, Tasmanian Outer Islands Association, Transcript of Evidence, 22 February 2000. 13. Council for Aboriginal Reconciliation, Corroboree 2000 : Towards Reconciliation. 14. Federal Court of Australia Decision, Edwina Shaw & Another v Charles Wolf & Others, [1998] 389 FAC (20 April 1998). 15. Flinders Island Council, Transcript of Evidence, 22 February 2000. 16. Gillies, Mrs Joy, Transcript of Evidence, 14 March 2000. 17. Government Media Statement, 12 October 1999. 18. Groom, Premier, Mr Ray, Second Reading Speech on Aboriginal Lands Bill 1995, 24 October 1995. 19. Herron MP, Senator John, Transcript of Meeting, 16 February 2000. 20. Hobart Quaker Peace and Justice Committee, Transcript of Evidence, 10 April 2000. 44 21. House, Mr Lyell, Forest, Transcript of Evidence, 14 March 2000. 22. Innes-Smith, Mr Peter, Submission to Legislative Council Select Committee on Aboriginal Lands, 23 January 2000. -
Figure B.9 Seasonal Distribution of ECSAS Other Alcid (Including Razorbill, Black Guillemot and Atlantic Puffin) Observations (2001 – 2017)
GXT GrandSPAN Marine Exploration Program 2018 Environmental Assessment Update Figure B.9 Seasonal Distribution of ECSAS Other Alcid (including Razorbill, Black Guillemot and Atlantic Puffin) Observations (2001 – 2017) GXT GrandSPAN Program (2014-2018) 2018 Environmental Assessment Update May 2018 Page 125 GXT GrandSPAN Marine Exploration Program 2018 Environmental Assessment Update Figure B.10 Seasonal Distribution of ECSAS Jaeger and Skua Observations (2001 – 2017) GXT GrandSPAN Program (2014-2018) 2018 Environmental Assessment Update May 2018 Page 126 GXT GrandSPAN Marine Exploration Program 2018 Environmental Assessment Update Figure B.11 Seasonal Distribution of ECSAS Northern Fulmar Observations (2001 – 2017) GXT GrandSPAN Program (2014-2018) 2018 Environmental Assessment Update May 2018 Page 127 GXT GrandSPAN Marine Exploration Program 2018 Environmental Assessment Update Figure B.12 Seasonal Distribution of ECSAS Shearwater Observations (2001 – 2017) GXT GrandSPAN Program (2014-2018) 2018 Environmental Assessment Update May 2018 Page 128 GXT GrandSPAN Marine Exploration Program 2018 Environmental Assessment Update Figure B.13 Seasonal Distribution of ECSAS Storm-petrel Observations (2001 – 2017) GXT GrandSPAN Program (2014-2018) 2018 Environmental Assessment Update May 2018 Page 129 GXT GrandSPAN Marine Exploration Program 2018 Environmental Assessment Update Figure B.14 Seasonal Distribution of ECSAS Waterfowl Observations (2001 – 2017) GXT GrandSPAN Program (2014-2018) 2018 Environmental Assessment Update May 2018