R-Trees: Introduction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tree-Combined Trie: a Compressed Data Structure for Fast IP Address Lookup
(IJACSA) International Journal of Advanced Computer Science and Applications, Vol. 6, No. 12, 2015 Tree-Combined Trie: A Compressed Data Structure for Fast IP Address Lookup Muhammad Tahir Shakil Ahmed Department of Computer Engineering, Department of Computer Engineering, Sir Syed University of Engineering and Technology, Sir Syed University of Engineering and Technology, Karachi Karachi Abstract—For meeting the requirements of the high-speed impact their forwarding capacity. In order to resolve two main Internet and satisfying the Internet users, building fast routers issues there are two possible solutions one is IPv6 IP with high-speed IP address lookup engine is inevitable. addressing scheme and second is Classless Inter-domain Regarding the unpredictable variations occurred in the Routing or CIDR. forwarding information during the time and space, the IP lookup algorithm should be able to customize itself with temporal and Finding a high-speed, memory-efficient and scalable IP spatial conditions. This paper proposes a new dynamic data address lookup method has been a great challenge especially structure for fast IP address lookup. This novel data structure is in the last decade (i.e. after introducing Classless Inter- a dynamic mixture of trees and tries which is called Tree- Domain Routing, CIDR, in 1994). In this paper, we will Combined Trie or simply TC-Trie. Binary sorted trees are more discuss only CIDR. In addition to these desirable features, advantageous than tries for representing a sparse population reconfigurability is also of great importance; true because while multibit tries have better performance than trees when a different points of this huge heterogeneous structure of population is dense. -

Balanced Trees Part One
Balanced Trees Part One Balanced Trees ● Balanced search trees are among the most useful and versatile data structures. ● Many programming languages ship with a balanced tree library. ● C++: std::map / std::set ● Java: TreeMap / TreeSet ● Many advanced data structures are layered on top of balanced trees. ● We’ll see several later in the quarter! Where We're Going ● B-Trees (Today) ● A simple type of balanced tree developed for block storage. ● Red/Black Trees (Today/Thursday) ● The canonical balanced binary search tree. ● Augmented Search Trees (Thursday) ● Adding extra information to balanced trees to supercharge the data structure. Outline for Today ● BST Review ● Refresher on basic BST concepts and runtimes. ● Overview of Red/Black Trees ● What we're building toward. ● B-Trees and 2-3-4 Trees ● Simple balanced trees, in depth. ● Intuiting Red/Black Trees ● A much better feel for red/black trees. A Quick BST Review Binary Search Trees ● A binary search tree is a binary tree with 9 the following properties: 5 13 ● Each node in the BST stores a key, and 1 6 10 14 optionally, some auxiliary information. 3 7 11 15 ● The key of every node in a BST is strictly greater than all keys 2 4 8 12 to its left and strictly smaller than all keys to its right. Binary Search Trees ● The height of a binary search tree is the 9 length of the longest path from the root to a 5 13 leaf, measured in the number of edges. 1 6 10 14 ● A tree with one node has height 0. -

Trees: Binary Search Trees
COMP2012H Spring 2014 Dekai Wu Binary Trees & Binary Search Trees (data structures for the dictionary ADT) Outline } Binary tree terminology } Tree traversals: preorder, inorder and postorder } Dictionary and binary search tree } Binary search tree operations } Search } min and max } Successor } Insertion } Deletion } Tree balancing issue COMP2012H (BST) Binary Tree Terminology } Go to the supplementary notes COMP2012H (BST) Linked Representation of Binary Trees } The degree of a node is the number of children it has. The degree of a tree is the maximum of its element degree. } In a binary tree, the tree degree is two data } Each node has two links left right } one to the left child of the node } one to the right child of the node Left child Right child } if no child node exists for a node, the link is set to NULL root 32 32 79 42 79 42 / 13 95 16 13 95 16 / / / / / / COMP2012H (BST) Binary Trees as Recursive Data Structures } A binary tree is either empty … Anchor or } Consists of a node called the root } Root points to two disjoint binary (sub)trees Inductive step left and right (sub)tree r left right subtree subtree COMP2012H (BST) Tree Traversal is Also Recursive (Preorder example) If the binary tree is empty then Anchor do nothing Else N: Visit the root, process data L: Traverse the left subtree Inductive/Recursive step R: Traverse the right subtree COMP2012H (BST) 3 Types of Tree Traversal } If the pointer to the node is not NULL: } Preorder: Node, Left subtree, Right subtree } Inorder: Left subtree, Node, Right subtree Inductive/Recursive step } Postorder: Left subtree, Right subtree, Node template <class T> void BinaryTree<T>::InOrder( void(*Visit)(BinaryTreeNode<T> *u), template<class T> BinaryTreeNode<T> *t) void BinaryTree<T>::PreOrder( {// Inorder traversal. -

Q-Tree: a Multi-Attribute Based Range Query Solution for Tele-Immersive Framework
Q-Tree: A Multi-Attribute Based Range Query Solution for Tele-Immersive Framework Md Ahsan Arefin, Md Yusuf Sarwar Uddin, Indranil Gupta, Klara Nahrstedt Department of Computer Science University of Illinois at Urbana Champaign Illinois, USA {marefin2, mduddin2, indy, klara}@illinois.edu Abstract given in a high level description which are transformed into multi-attribute composite range queries. Some of the Users and administrators of large distributed systems are examples include “which site is highly congested?”, “which frequently in need of monitoring and management of its components are not working properly?” etc. To answer the various components, data items and resources. Though there first one, the query is transformed into a multi-attribute exist several distributed query and aggregation systems, the composite range query with constrains (range of values) on clustered structure of tele-immersive interactive frameworks CPU utilization, memory overhead, stream rate, bandwidth and their time-sensitive nature and application requirements utilization, delay and packet loss rate. The later one can represent a new class of systems which poses different chal- be answered by constructing a multi-attribute range query lenges on this distributed search. Multi-attribute composite with constrains on static and dynamic characteristics of range queries are one of the key features in this class. those components. Queries can also be made by defining Queries are given in high level descriptions and then trans- different multi-attribute ranges explicitly. Another mention- formed into multi-attribute composite range queries. Design- able property of such systems is that the number of site is ing such a query engine with minimum traffic overhead, low limited due to limited display space and limited interactions, service latency, and with static and dynamic nature of large but the number of data items to store and manage can datasets, is a challenging task. -

Parameter-Free Locality Sensitive Hashing for Spherical Range Reporting˚
Parameter-free Locality Sensitive Hashing for Spherical Range Reporting˚ Thomas D. Ahle, Martin Aumüller, and Rasmus Pagh IT University of Copenhagen, Denmark, {thdy, maau, pagh}@itu.dk July 21, 2016 Abstract We present a data structure for spherical range reporting on a point set S, i.e., reporting all points in S that lie within radius r of a given query point q (with a small probability of error). Our solution builds upon the Locality-Sensitive Hashing (LSH) framework of Indyk and Motwani, which represents the asymptotically best solutions to near neighbor problems in high dimensions. While traditional LSH data structures have several parameters whose optimal values depend on the distance distribution from q to the points of S (and in particular on the number of points to report), our data structure is essentially parameter-free and only takes as parameter the space the user is willing to allocate. Nevertheless, its expected query time basically matches that of an LSH data structure whose parameters have been optimally chosen for the data and query in question under the given space constraints. In particular, our data structure provides a smooth trade-off between hard queries (typically addressed by standard LSH parameter settings) and easy queries such as those where the number of points to report is a constant fraction of S, or where almost all points in S are far away from the query point. In contrast, known data structures fix LSH parameters based on certain parameters of the input alone. The algorithm has expected query time bounded by Optpn{tqρq, where t is the number of points to report and ρ P p0; 1q depends on the data distribution and the strength of the LSH family used. -
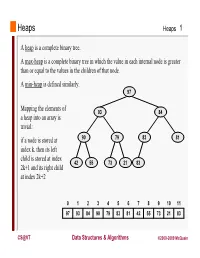
Heaps a Heap Is a Complete Binary Tree. a Max-Heap Is A
Heaps Heaps 1 A heap is a complete binary tree. A max-heap is a complete binary tree in which the value in each internal node is greater than or equal to the values in the children of that node. A min-heap is defined similarly. 97 Mapping the elements of 93 84 a heap into an array is trivial: if a node is stored at 90 79 83 81 index k, then its left child is stored at index 42 55 73 21 83 2k+1 and its right child at index 2k+2 01234567891011 97 93 84 90 79 83 81 42 55 73 21 83 CS@VT Data Structures & Algorithms ©2000-2009 McQuain Building a Heap Heaps 2 The fact that a heap is a complete binary tree allows it to be efficiently represented using a simple array. Given an array of N values, a heap containing those values can be built, in situ, by simply “sifting” each internal node down to its proper location: - start with the last 73 73 internal node * - swap the current 74 81 74 * 93 internal node with its larger child, if 79 90 93 79 90 81 necessary - then follow the swapped node down 73 * 93 - continue until all * internal nodes are 90 93 90 73 done 79 74 81 79 74 81 CS@VT Data Structures & Algorithms ©2000-2009 McQuain Heap Class Interface Heaps 3 We will consider a somewhat minimal maxheap class: public class BinaryHeap<T extends Comparable<? super T>> { private static final int DEFCAP = 10; // default array size private int size; // # elems in array private T [] elems; // array of elems public BinaryHeap() { . -
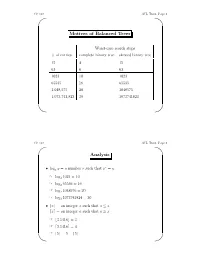
AVL Trees, Page 1 ' $
CS 310 AVL Trees, Page 1 ' $ Motives of Balanced Trees Worst-case search steps #ofentries complete binary tree skewed binary tree 15 4 15 63 6 63 1023 10 1023 65535 16 65535 1,048,575 20 1048575 1,073,741,823 30 1073741823 & % CS 310 AVL Trees, Page 2 ' $ Analysis r • logx y =anumberr such that x = y ☞ log2 1024 = 10 ☞ log2 65536 = 16 ☞ log2 1048576 = 20 ☞ log2 1073741824 = 30 •bxc = an integer a such that a ≤ x dxe = an integer a such that a ≥ x ☞ b3.1416c =3 ☞ d3.1416e =4 ☞ b5c =5=d5e & % CS 310 AVL Trees, Page 3 ' $ • If a binary search tree of N nodes happens to be complete, then a search in the tree requires at most b c log2 N +1 steps. • Big-O notation: We say f(x)=O(g(x)) if f(x)isbounded by c · g(x), where c is a constant, for sufficiently large x. ☞ x2 +11x − 30 = O(x2) (consider c =2,x ≥ 6) ☞ x100 + x50 +2x = O(2x) • If a BST happens to be complete, then a search in the tree requires O(log2 N)steps. • In a linked list of N nodes, a search requires O(N)steps. & % CS 310 AVL Trees, Page 4 ' $ AVL Trees • A balanced binary search tree structure proposed by Adelson, Velksii and Landis in 1962. • In an AVL tree of N nodes, every searching, deletion or insertion operation requires only O(log2 N)steps. • AVL tree searching is exactly the same as that of the binary search tree. & % CS 310 AVL Trees, Page 5 ' $ Definition • An empty tree is balanced. -

Binary Search Tree
ADT Binary Search Tree! Ellen Walker! CPSC 201 Data Structures! Hiram College! Binary Search Tree! •" Value-based storage of information! –" Data is stored in order! –" Data can be retrieved by value efficiently! •" Is a binary tree! –" Everything in left subtree is < root! –" Everything in right subtree is >root! –" Both left and right subtrees are also BST#s! Operations on BST! •" Some can be inherited from binary tree! –" Constructor (for empty tree)! –" Inorder, Preorder, and Postorder traversal! •" Some must be defined ! –" Insert item! –" Delete item! –" Retrieve item! The Node<E> Class! •" Just as for a linked list, a node consists of a data part and links to successor nodes! •" The data part is a reference to type E! •" A binary tree node must have links to both its left and right subtrees! The BinaryTree<E> Class! The BinaryTree<E> Class (continued)! Overview of a Binary Search Tree! •" Binary search tree definition! –" A set of nodes T is a binary search tree if either of the following is true! •" T is empty! •" Its root has two subtrees such that each is a binary search tree and the value in the root is greater than all values of the left subtree but less than all values in the right subtree! Overview of a Binary Search Tree (continued)! Searching a Binary Tree! Class TreeSet and Interface Search Tree! BinarySearchTree Class! BST Algorithms! •" Search! •" Insert! •" Delete! •" Print values in order! –" We already know this, it#s inorder traversal! –" That#s why it#s called “in order”! Searching the Binary Tree! •" If the tree is -

7. Hierarchies & Trees Visualizing Topological Relations
7. Hierarchies & Trees Visualizing topological relations Vorlesung „Informationsvisualisierung” Prof. Dr. Andreas Butz, WS 2011/12 Konzept und Basis für Folien: Thorsten Büring LMU München – Medieninformatik – Andreas Butz – Informationsvisualisierung – WS2011/12 Folie 1 Outline • Hierarchical data and tree representations • 2D Node-link diagrams – Hyperbolic Tree Browser – SpaceTree – Cheops – Degree of interest tree – 3D Node-link diagrams • Enclosure – Treemap – Ordererd Treemaps – Various examples – Voronoi treemap – 3D Treemaps • Circular visualizations • Space-filling node-link diagram LMU München – Medieninformatik – Andreas Butz – Informationsvisualisierung – WS2011/12 Folie 2 Hierarchical Data • Card et al. 1999: data repository in which data cases are related to subcases • Many data collections have an inherent hierarchical organization – Organizational Charts – Websites (approximately hierarchical) Yee et al. 2001 – File system – Family tree – OO programming • Hierarchies are usually represented as tree visual structures • Trees tend to be easier to lay out and interpret than networks (e.g. no cycles) • But: as shown in the example, networks may in some cases be visualized as a tree LMU München – Medieninformatik – Andreas Butz – Informationsvisualisierung – WS2011/12 Folie 3 Tree Representations • Two kinds of representations • Node-link diagram (see previous lecture): represent connections as edges between vertices (data cases) http://www.icann.org • Enclosure: space-filling approaches by visually nesting the hierarchy LMU -

6.172 Lecture 19 : Cache-Oblivious B-Tree (Tokudb)
How TokuDB Fractal TreeTM Indexes Work Bradley C. Kuszmaul Guest Lecture in MIT 6.172 Performance Engineering, 18 November 2010. 6.172 —How Fractal Trees Work 1 My Background • I’m an MIT alum: MIT Degrees = 2 × S.B + S.M. + Ph.D. • I was a principal architect of the Connection Machine CM-5 super computer at Thinking Machines. • I was Assistant Professor at Yale. • I was Akamai working on network mapping and billing. • I am research faculty in the SuperTech group, working with Charles. 6.172 —How Fractal Trees Work 2 Tokutek A few years ago I started collaborating with Michael Bender and Martin Farach-Colton on how to store data on disk to achieve high performance. We started Tokutek to commercialize the research. 6.172 —How Fractal Trees Work 3 I/O is a Big Bottleneck Sensor Query Systems include Sensor Disk Query sensors and Sensor storage, and Query want to perform Millions of data elements arrive queries on per second Query recently arrived data using indexes. recent data. Sensor 6.172 —How Fractal Trees Work 4 The Data Indexing Problem • Data arrives in one order (say, sorted by the time of the observation). • Data is queried in another order (say, by URL or location). Sensor Query Sensor Disk Query Sensor Query Millions of data elements arrive per second Query recently arrived data using indexes. Sensor 6.172 —How Fractal Trees Work 5 Why Not Simply Sort? • This is what data Data Sorted by Time warehouses do. • The problem is that you Sort must wait to sort the data before querying it: Data Sorted by URL typically an overnight delay. -

On Range Searching with Semialgebraic Sets*
Discrete Comput Geom 11:393~,18 (1994) GeometryDiscrete & Computational 1994 Springer-Verlag New York Inc. On Range Searching with Semialgebraic Sets* P. K. Agarwal 1 and J. Matou~ek 2 1 Computer Science Department, Duke University, Durham, NC 27706, USA 2 Katedra Aplikovan6 Matamatiky, Universita Karlova, 118 00 Praka 1, Czech Republic and Institut fiir Informatik, Freie Universit~it Berlin, Arnirnallee 2-6, D-14195 Berlin, Germany matou~ek@cspguk 11.bitnet Abstract. Let P be a set of n points in ~d (where d is a small fixed positive integer), and let F be a collection of subsets of ~d, each of which is defined by a constant number of bounded degree polynomial inequalities. We consider the following F-range searching problem: Given P, build a data structure for efficient answering of queries of the form, "Given a 7 ~ F, count (or report) the points of P lying in 7." Generalizing the simplex range searching techniques, we give a solution with nearly linear space and preprocessing time and with O(n 1- x/b+~) query time, where d < b < 2d - 3 and ~ > 0 is an arbitrarily small constant. The acutal value of b is related to the problem of partitioning arrangements of algebraic surfaces into cells with a constant description complexity. We present some of the applications of F-range searching problem, including improved ray shooting among triangles in ~3 1. Introduction Let F be a family of subsets of the d-dimensional space ~d (d is a small constant) such that each y e F can be described by some fixed number of real parameters (for example, F can be the set of balls, or the set of all intersections of two ellipsoids, * Part of the work by P. -

New Data Structures for Orthogonal Range Searching
Alcom-FT Technical Report Series ALCOMFT-TR-01-35 New Data Structures for Orthogonal Range Searching Stephen Alstrup∗ The IT University of Copenhagen Glentevej 67, DK-2400, Denmark [email protected] Gerth Stølting Brodal† BRICS ‡ Department of Computer Science University of Aarhus Ny Munkegade, DK-8000 Arhus˚ C, Denmark [email protected] Theis Rauhe§ The IT University of Copenhagen Glentevej 67, DK-2400, Denmark [email protected] 5th April 2001 Abstract We present new general techniques for static orthogonal range searching problems in two and higher dimensions. For the general range reporting problem in R3, we achieve ε query time O(log n + k) using space O(n log1+ n), where n denotes the number of stored points and k the number of points to be reported. For the range reporting problem on ε an n n grid, we achieve query time O(log log n + k) using space O(n log n). For the two-dimensional× semi-group range sum problem we achieve query time O(log n) using space O(n log n). ∗Partially supported by a grant from Direktør Ib Henriksens Fond. †Partially supported by the IST Programme of the EU under contract number IST-1999-14186 (ALCOM- FT). ‡Basic Research in Computer Science, Center of the Danish National Research Foundation. §Partially supported by a grant from Direktør Ib Henriksens Fond. Part of the work was done while the author was at BRICS. 1 1 Introduction Let P be a finite set of points in Rd and Q a query range in Rd. Range searching is the problem of answering various types of queries about the set of points which are contained within the query range, i.e., the point set P Q.