Fifty-Million-Year-Old Polymorphism at an Immunoglobulin Variable Region Gene Locus in the Rabbit Evolutionary Lineage
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Las Vegas Channel Lineup
Las Vegas Channel Lineup PrismTM TV 222 Bloomberg Interactive Channels 5145 Tropicales 225 The Weather Channel 90 Interactive Dashboard 5146 Mexicana 2 City of Las Vegas Television 230 C-SPAN 92 Interactive Games 5147 Romances 3 NBC 231 C-SPAN2 4 Clark County Television 251 TLC Digital Music Channels PrismTM Complete 5 FOX 255 Travel Channel 5101 Hit List TM 6 FOX 5 Weather 24/7 265 National Geographic Channel 5102 Hip Hop & R&B Includes Prism TV Package channels, plus 7 Universal Sports 271 History 5103 Mix Tape 132 American Life 8 CBS 303 Disney Channel 5104 Dance/Electronica 149 G4 9 LATV 314 Nickelodeon 5105 Rap (uncensored) 153 Chiller 10 PBS 326 Cartoon Network 5106 Hip Hop Classics 157 TV One 11 V-Me 327 Boomerang 5107 Throwback Jamz 161 Sleuth 12 PBS Create 337 Sprout 5108 R&B Classics 173 GSN 13 ABC 361 Lifetime Television 5109 R&B Soul 188 BBC America 14 Mexicanal 362 Lifetime Movie Network 5110 Gospel 189 Current TV 15 Univision 364 Lifetime Real Women 5111 Reggae 195 ION 17 Telefutura 368 Oxygen 5112 Classic Rock 253 Animal Planet 18 QVC 420 QVC 5113 Retro Rock 257 Oprah Winfrey Network 19 Home Shopping Network 422 Home Shopping Network 5114 Rock 258 Science Channel 21 My Network TV 424 ShopNBC 5115 Metal (uncensored) 259 Military Channel 25 Vegas TV 428 Jewelry Television 5116 Alternative (uncensored) 260 ID 27 ESPN 451 HGTV 5117 Classic Alternative 272 Biography 28 ESPN2 453 Food Network 5118 Adult Alternative (uncensored) 274 History International 33 CW 503 MTV 5120 Soft Rock 305 Disney XD 39 Telemundo 519 VH1 5121 Pop Hits 315 Nick Too 109 TNT 526 CMT 5122 90s 316 Nicktoons 113 TBS 560 Trinity Broadcasting Network 5123 80s 320 Nick Jr. -

VH1-6,8,10 Vent Hoods 8504054 Rev B 09/00
OWNERS INSTRUCTIONS THESE INSTRUCTIONS TJERNLUND PRODUCTS, INC. MUST REMAIN 1601 Ninth Street • White Bear Lake, MN 55110-6794 WITH EQUIPMENT PHONE (651) 426-2993 • (800) 255-4208 • FAX (651) 426-9547 DO NOT DESTROY Visit our web site • www.tjernlund.com MODEL VH1 SERIES VENT HOODS 6”, 8”, 10” The VH1-8 and VH1-10 do not have a secondary heat shield. A thimble will have to be added to maintain clearances to combustibles if penetrating a combustible wall with these models. 1. Terminate the vent system so that proper minimum clearances are maintained as cited in the latest edition of the National Fuel Gas Code (NFPA # 54) and the latest edition of NFPA #211, or as follows: • Not be less than 7 feet above grade when located adjacent to public walk ways • At least 3 feet above any forced air inlet located within 10 feet • At least 4 feet below, 4 feet horizontally from or 1 foot above any door, window or gravity air inlet into any building • At least 12 inches above grade. THE MANUFACTURER ALSO RECOMMMENDS: • The vent terminal shall also not be installed closer than 3 feet from the inside corner of an L-shaped structure • Not less than 2 feet from an adjacent building • Flue gases are not directed so as to jeopardize people, overheat combustible structures or enter buildings 2. Check vent pipe system for leakage. All vent pipe joints on “positive” side of power venter must be sealed. 3. A vent system incorporating a Tjernlund VH1 Series vent hood should not exceed 550o F. -

Xfinity Channel Lineup
Channel Lineup 1-800-XFINITY | xfinity.com SARASOTA, MANATEE, VENICE, VENICE SOUTH, AND NORTH PORT Legend Effective: April 1, 2016 LIMITED BASIC 26 A&E 172 UP 183 QUBO 738 SPORTSMAN CHANNEL 1 includes Music Choice 27 HLN 179 GSN 239 JLTV 739 NHL NETWORK 2 ION (WXPX) 29 ESPN 244 INSP 242 TBN 741 NFL REDZONE <2> 3 PBS (WEDU SARASOTA & VENICE) 30 ESPN2 42 BLOOMBERG 245 PIVOT 742 BTN 208 LIVE WELL (WSNN) 31 THE WEATHER CHANNEL 719 HALLMARK MOVIES & MYSTERIES 246 BABYFIRST TV AMERICAS 744 ESPNU 5 HALLMARK CHANNEL 32 CNN 728 FXX (ENGLISH) 746 MAV TV 6 SUNCOAST NEWS (WSNN) 33 MTV 745 SEC NETWORK 247 THE WORD NETWORK 747 WFN 7 ABC (WWSB) 34 USA 768-769 SEC NETWORK (OVERFLOW) 248 DAYSTAR 762 CSN - CHICAGO 8 NBC (WFLA) 35 BET 249 JUCE 764 PAC 12 9 THE CW (WTOG) 36 LIFETIME DIGITAL PREFERRED 250 SMILE OF A CHILD 765 CSN - NEW ENGLAND 10 CBS (WTSP) 37 FOOD NETWORK 1 includes Digital Starter 255 OVATION 766 ESPN GOAL LINE <14> 11 MY NETWORK TV (WTTA) 38 FOX SPORTS SUN 57 SPIKE 257 RLTV 785 SNY 12 IND (WMOR) 39 CNBC 95 POP 261 FAMILYNET 47, 146 CMT 13 FOX (WTVT) 40 DISCOVERY CHANNEL 101 WEATHERSCAN 271 NASA TV 14 QVC 41 HGTV 102, 722 ESPNEWS 279 MLB NETWORK MUSIC CHOICE <3> 15 UNIVISION (WVEA) 44 ANIMAL PLANET 108 NAT GEO WILD 281 FX MOVIE CHANNEL 801-850 MUSIC CHOICE 17 PBS (WEDU VENICE SOUTH) 45 TLC 110 SCIENCE 613 GALAVISION 17 ABC (WFTS SARASOTA) 46 E! 112 AMERICAN HEROES 636 NBC UNIVERSO ON DEMAND TUNE-INS 18 C-SPAN 48 FOX SPORTS ONE 113 DESTINATION AMERICA 667 UNIVISION DEPORTES <5> 19 LOCAL GOVT (SARASOTA VENICE & 49 GOLF CHANNEL 121 DIY NETWORK 721 TV GAMES 1 includes Limited Basic VENICE SOUTH) 50 VH1 122 COOKING CHANNEL 734 NBA TV 1, 199 ON DEMAND (MAIN MENU) 19 LOCAL EDUCATION (MANATEE) 51 FX 127 SMITHSONIAN CHANNEL 735 CBS SPORTS NETWORK 194 MOVIES ON DEMAND 20 LOCAL GOVT (MANATEE) 55 FREEFORM 129 NICKTOONS 738 SPORTSMAN CHANNEL 299 FREE MOVIES ON DEMAND 20 LOCAL EDUCATION (SARASOTA, 56 AMC 130 DISCOVERY FAMILY CHANNEL 739 NHL NETWORK 300 HBO ON DEMAND VENICE & VENICE SOUTH) 58 OWN 131 NICK JR. -

Hawaiian Telcom TV Channel Packages
Hawaiian Telcom TV 604 Stingray Everything 80’s ADVANTAGE PLUS 1003 FOX-KHON HD 1208 BET HD 1712 Pets.TV 525 Thriller Max 605 Stingray Nothin but 90’s 21 NHK World 1004 ABC-KITV HD 1209 VH1 HD MOVIE VARIETY PACK 526 Movie MAX Channel Packages 606 Stingray Jukebox Oldies 22 Arirang TV 1005 KFVE (Independent) HD 1226 Lifetime HD 380 Sony Movie Channel 527 Latino MAX 607 Stingray Groove (Disco & Funk) 23 KBS World 1006 KBFD (Korean) HD 1227 Lifetime Movie Network HD 381 EPIX 1401 STARZ (East) HD ADVANTAGE 125 TNT 608 Stingray Maximum Party 24 TVK1 1007 CBS-KGMB HD 1229 Oxygen HD 382 EPIX 2 1402 STARZ (West) HD 1 Video On Demand Previews 126 truTV 609 Stingray Dance Clubbin’ 25 TVK2 1008 NBC-KHNL HD 1230 WE tv HD 387 STARZ ENCORE 1405 STARZ Kids & Family HD 2 CW-KHON 127 TV Land 610 Stingray The Spa 28 NTD TV 1009 QVC HD 1231 Food Network HD 388 STARZ ENCORE Black 1407 STARZ Comedy HD 3 FOX-KHON 128 Hallmark Channel 611 Stingray Classic Rock 29 MYX TV (Filipino) 1011 PBS-KHET HD 1232 HGTV HD 389 STARZ ENCORE Suspense 1409 STARZ Edge HD 4 ABC-KITV 129 A&E 612 Stingray Rock 30 Mnet 1017 Jewelry TV HD 1233 Destination America HD 390 STARZ ENCORE Family 1451 Showtime HD 5 KFVE (Independent) 130 National Geographic Channel 613 Stingray Alt Rock Classics 31 PAC-12 National 1027 KPXO ION HD 1234 DIY Network HD 391 STARZ ENCORE Action 1452 Showtime East HD 6 KBFD (Korean) 131 Discovery Channel 614 Stingray Rock Alternative 32 PAC-12 Arizona 1069 TWC SportsNet HD 1235 Cooking Channel HD 392 STARZ ENCORE Classic 1453 Showtime - SHO2 HD 7 CBS-KGMB 132 -

DIGITAL TV LINEUP SD HD Lifeline SD HD SD HD SD HD SD HD 2 NBC-KSNC/KSNW - HD 500 Velocity - HD 58 558 Science Channel 90 590 Disney Channel 143 Natoma School Ch
DIGITAL TV LINEUP SD HD Lifeline SD HD SD HD SD HD SD HD 2 NBC-KSNC/KSNW - HD 500 Velocity - HD 58 558 Science Channel 90 590 Disney Channel 143 Natoma School Ch. 3 The Weather Ch. - HD 25 525 ESPN 59 Game Show Network 92 Cartoon Network 144 Victoria School Ch. 4 FOX - KSAS-HD 27 527 ESPNEWS 60 A&E Network - HD 93 593 Discovery Family Ch. 145 Russell School Ch. 5 PBS - Create TV 28 528 ESPNU 61 History Channel - HD 94 Nickelodeon 625 MTV Live - HD 6 GCI Local 29 529 ESPN2 62 Viceland 95 Nick Jr. 7 CBS - KBSH/KWCH -HD 30 530 Big Ten 64 FYI 99 Boomerang 8 MeTV - KAKE 31 NFL Network - HD 65 TV Land 110 FX Network 9 PBS - KOOD - HD 32 532 Fox Sports 1 66 Paramount Network 112 Lifetime Movie Net. - HD 10 ABC - KAKE - HD 33 Fox Sports - Midwest 67 Comedy Central 113 Turner Class. Movies 11 EWTN 40 540 CNN 68 BET 114 Hallmark Movies & Myst 12 TBN 41 541 CNN Headline News 69 Lifetime Telev. - HD 115 FXX Network 13 C-SPAN 42 Fox News Ch - HD 70 Lifetime Real Women 120 VH1 14 C-SPAN2 43 Fox Business News 71 Hallmark Channel 123 MTV 15 C-SPAN3 50 550 National Geographic 72 572 HGTV 124 MTV2 additional for SPANISH 16 Stadium Ch-KMTW 51 551 Discovery Channel 73 573 Food Network 128 MTV-U 180 Discovery Familia 17 Antenna TV-KSCW 52 The Oprah Winfrey Net. 181 Discovery en Espanol 18 PBS Kids 53 553 Animal Planet 78 578 TNT 130 CMT 20 Stormtracker-KWCH 54 American Heroes 79 579 TBS 132 GAC 182 History en Espanol 21 My TV - KMTW - HD 55 555 The Learning Channel 80 truTV 140 QVC 183 CNN en Espanol 22 CW - KSCW- HD 56 BBC America 83 583 Investigation Disc. -
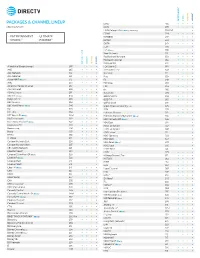
Packages & Channel Lineup
™ ™ ENTERTAINMENT CHOICE ULTIMATE PREMIER PACKAGES & CHANNEL LINEUP ESNE3 456 • • • • Effective 6/17/21 ESPN 206 • • • • ESPN College Extra2 (c only) (Games only) 788-798 • ESPN2 209 • • • • • ENTERTAINMENT • ULTIMATE ESPNEWS 207 • • • • CHOICE™ • PREMIER™ ESPNU 208 • • • EWTN 370 • • • • FLIX® 556 • FM2 (c only) 386 • • Food Network 231 • • • • ™ ™ Fox Business Network 359 • • • • Fox News Channel 360 • • • • ENTERTAINMENT CHOICE ULTIMATE PREMIER FOX Sports 1 219 • • • • A Wealth of Entertainment 387 • • • FOX Sports 2 618 • • A&E 265 • • • • Free Speech TV3 348 • • • • ACC Network 612 • • • Freeform 311 • • • • AccuWeather 361 • • • • Fuse 339 • • • ActionMAX2 (c only) 519 • FX 248 • • • • AMC 254 • • • • FX Movie 258 • • American Heroes Channel 287 • • FXX 259 • • • • Animal Planet 282 • • • • fyi, 266 • • ASPiRE2 (HD only) 381 • • Galavisión 404 • • • • AXS TV2 (HD only) 340 • • • • GEB America3 363 • • • • BabyFirst TV3 293 • • • • GOD TV3 365 • • • • BBC America 264 • • • • Golf Channel 218 • • 2 c BBC World News ( only) 346 • • Great American Country (GAC) 326 • • BET 329 • • • • GSN 233 • • • BET HER 330 • • Hallmark Channel 312 • • • • BET West HD2 (c only) 329-1 2 • • • • Hallmark Movies & Mysteries (c only) 565 • • Big Ten Network 610 2 • • • HBO Comedy HD (c only) 506 • 2 Black News Channel (c only) 342 • • • • HBO East 501 • Bloomberg TV 353 • • • • HBO Family East 507 • Boomerang 298 • • • • HBO Family West 508 • Bravo 237 • • • • HBO Latino3 511 • BYUtv 374 • • • • HBO Signature 503 • C-SPAN2 351 • • • • HBO West 504 • -

XFINITY® TV Channel Lineup
XFINITY® TV Channel Lineup Somerville, MA C-103 | 05.13 51 NESN 837 A&E HD 852 Comcast SportsNet HD Limited Basic 52 Comcast SportsNet 841 Fox News HD 854 Food Network HD 54 BET 842 CNN HD 855 Spike TV HD 2 WGBH-2 (PBS) / HD 802 55 Spike TV 854 Food Network HD 858 Comedy Central HD 3 Public Access 57 Bravo 859 AMC HD 859 AMC HD 4 WBZ-4 (CBS) / HD 804 59 AMC 863 Animal Planet HD 860 Cartoon Network HD 5 WCVB-5 (ABC) / HD 805 60 Cartoon Network 872 History HD 862 Syfy HD 6 NECN 61 Comedy Central 905 BET HD 863 Animal Planet HD 7 WHDH-7 (NBC) / HD 807 62 Syfy 906 HSN HD 865 NBC Sports Network HD 8 HSN 63 Animal Planet 907 Hallmark HD 867 TLC HD 9 WBPX-68 (ION) / HD 803 64 TV Land 910 H2 HD 872 History HD 10 WWDP-DT 66 History 901 MSNBC HD 67 Travel Channel 902 truTV HD 12 WLVI-56 (CW) / HD 808 13 WFXT-25 (FOX) / HD 806 69 Golf Channel Digital Starter 905 BET HD 14 WSBK myTV38 (MyTV) / 186 truTV (Includes Limited Basic and 906 HSN HD HD 814 208 Hallmark Channel Expanded Basic) 907 Hallmark HD 15 Educational Access 234 Inspirational Network 908 GMC HD 16 WGBX-44 (PBS) / HD 801 238 EWTN 909 Investigation Discovery HD 251 MSNBC 1 On Demand 910 H2 HD 17 WUNI-27 (UNI) / HD 816 42/246 Bloomberg Television 18 WBIN (IND) / HD 811 270 Lifetime Movie Network 916 Bloomberg Television HD 284 Fox Business Network 182 TV Guide Entertainment 920 BBC America HD 19 WNEU-60 (Telemundo) / 199 Hallmark Movie Channel HD 815 200 MoviePlex 20 WMFP-62 (IND) / HD 813 Family Tier 211 style. -

Channel Guide Packages and Channels Subject to Change 855.269.3429
Channel Guide Packages and channels subject to change 855.269.3429 www.southcentralconnect.net 1 Create 164 ESPN Classic 506 Nick Too 859 Movie Channel 2 PBS 165 NFL Red Zone 507 Nicktoons 860 Movie Channel Xtra 3 PBS Kids 166 ESPN Goal Line 508 Teen Nick 900 GAC 4 KARK NBC 190 Event PPV HD 509 UP 901 CMT Music 5 La 191 Event PPV SD 510 Boomerang 902 Nick Music 6 Grit 200 USA 511 Cartoon Network 903 MTV Classic 7 KATV ABC 201 TBS 512 Discovery Family 904 MTV2 8 Comet 202 TNT 600 Discovery Channel 905 BET Jams 9 Charge! 203 TruTV 601 Animal Planet 906 BET Soul 10 TBD 204 Bravo 602 National Geographic 920 Stingray Hit List 11 KTHV CBS 205 SyFy 606 Travel 921 Stingray Pop Adult 12 Antenna TV 206 AMC 607 Velocity 922 Stingray Standards 13 Justice 207 A&E 700 Lifetime 923 Stingray Jukebox Oldies 14 Quest 208 History Channel 701 Lifetime Movie Network 924 Stingray Flashback 70s 16 KLRT Fox 209 WGN 702 WE 925 Stingray Everything 80s 17 Escape 210 Hallmark Channel 703 OWN 926 Stingray Nothin But 90s 23 QVC 211 BBC America 704 Oxygen 927 Stingray Maximum Party 30 Telemundo 212 GSN 800 FX Movie 928 Stingray Groove Disco & Funk 31 Univision 215 Discovery Life 801 IFC 929 Stingray Dance Clubbin 32 Buzzr 220 Logo 802 Sundance 930 Stingray Holiday Hits 33 Evine 221 Spike 803 Turner Classic Movies 931 Stingray Classic Rock 35 NASA 222 TV Land 804 MGM 932 Stingray Rock Alternative 36 Daystar 223 Comedy Central 820 HBO 933 Stingray Rock 38 KASN CW 224 FX 821 HBO 2 934 Stingray Heavy Metal 42 MyTV 225 FXX 822 HBO Comedy 935 Stingray Alt Rock Classics 43 -

Channel Directory Channel Directory
Name Number Call Letters Name Number Call Letters Name Number Call Letters Fox News Channel FNC 210 PBS KIDS Sprout SPROUT 337 Cinemax MAX 832 Cleveland Fox Reality Channel REAL 130 qubo qubo 328 Cinemax - West MAX-W 833 Fox Soccer Channel ** FSC 654 QVC QVC 197 Encore ENC 932 Fox Sports en Español ** FSE 655 QVC QVC 420 Encore - West ENC-W 933 FSN Arizona ** FSAZ 762 Recorded TV Channel DVR 9999 Encore Action ENCACT 936 Channel Directory FSN Detroit ** FSD 737 Sci Fi Channel SCIFI 151 Encore Drama ENCDRA 938 BY CHANNEL NAME FSN Florida ** FSFL 720 Science Channel SCI 258 Encore Love ENCLOV 934 FSN Midwest ** FSMW 748 ShopNBC SHPNBC 424 Encore Mystery ENCMYS 935 FSN North ** FSN 744 SiTV SiTV 194 Encore Wam WAM 939 Name Call Letters Number FSN Northwest ** FSNW 764 Sleuth SLEUTH 161 Encore Westerns ENCWES 937 FSN Ohio-Cincinnati ** FSOHCI 732 Smile of a Child SMILE 340 FLIX FLIX 890 FSN Ohio-Cleveland ** FSOHCL 734 SOAPnet SOAP 365 HBO HBO 802 LOCAL LISTINGS FSN Pittsburgh ** FSP 730 SOAPnet - West SOAP-W 366 HBO - West HBO-W 803 FSN Prime Ticket ** FSPT 774 Speed Channel ** SPEED 652 HBO Comedy HBOCOM 808 HSN HSN 18 FSN Rocky Mountain ** FSRM 760 Spike TV SPKE 145 HBO Family HBOFAM 806 WBNX-55 (THE CW) WBNX 55 FSN South ** FSS 724 Spike TV - West SPKE-W 146 HBO Latino HBOLAT 810 WDLI-17 (TBN) WDLI 17 FSN Southwest ** FSSW 753 Sports Time Ohio STO 735 HBO Signature HBOSIG 807 WEAO-49 (PBS) WEAO 49 FSN West ** FSW 772 SportsNet New York ** SNNY 704 HBO Zone HBOZNE 809 WEWS-5 (ABC) WEWS 5 Fuel FUEL 536 SportSouth ** SPTSO 729 HBO2 HBO2 -

Digital Favorites Channel Lineup
Two City Plaza CUSTOMER SERVICE: 1-800-355-5668 Digital Favorites Channel Lineup 2 PBS2 - WPBT 47 SEC Network Overflow 85 BET 3 NBC 5 - WPTV 48 CNBC 86 MTV 5 Univision 49 CNN 87 MTV2 7 Create TV WPBT 50 HLN 88 NickMusic 8 Telemundo 51 BBC America 89 BET Jams 9 UniMas 52 FOX News 90 VH1 11 WeatherNation WPEC 53 MSNBC 91 MTV Classic 12 CBS 12 - WPEC 54 C-SPAN 92 BET Soul 13 ABC 25 - WPBF 55 C-SPAN 2 93 CMT 14 FOX 29 - WFLX 56 C-SPAN 3 94 CMT Music 15 CW 34 - WTVX 57 Discovery Channel 95 A&E Network 16 PBS 42 - WPBT 58 OWN 96 Oxygen 17 CTN 61 - WFGC 59 TLC 97 DIY Network 18 ION 67 - WPXP 60 The History Channel 98 Cooking Channel 19 My 15 WTCN-TV 61 Animal Planet 99 LOGO 20 PBC TV Channel 20 62 Freeform 100 Hallmark Channel 23 PBS 17 - WLRN 63 Disney XD 101 Lifetime Movie Network 24 Comet WPEC 64 Disney 102 Lifetime Real Women 25 The Weather Channel 65 Nickelodeon 103 WE 26 Lifetime 66 TeenNick 104 Paramount Network 27 AccuWeather 67 Nicktoons 105 GAC Family 28 One America News 68 Nick 2 106 BET Her 29 Comedy Central 69 Nick Jr 111 Investigation Discovery 30 Bally Sports Florida 70 TV Land 113 Enrich 31 Bally Sports Sun 72 E! 114 BET Gospel 32 ESPN 73 Food Network 115 BBC World 33 ESPN2 74 HGTV 117 ShopHQ 34 ESPNEWS 75 Disney Junior 118 EWTN 35 ESPN U 76 Travel Channel 119 The Olympic Channel 37 NHL Network 77 Bravo 120 FOX Business Network 38 NBC Sports Network 78 SyFy 121 Newsmax TV 39 The Golf Channel 79 FX 125 MTVU 40 NFL Network 80 TNT 205 FXX 41 MLB Network 81 TBS 207 Tennis Channel 43 ACC Network 82 USA Network 208 Big Ten Network -

Alphabetical Channel Listing
EXPANDED BASIC Alphabetical Channel Listing 129 A&E 64 Eternal Word 101 Nick 2 236 Wealth TV 604 JUKEBOX OLDIES 761 A&E HD* 231 Food Network 104 Nick Jr. 763 Wealth TV HD* 650 KID’S STUFF 122 ABC Family 755 Food Network HD* 100 Nickelodeon/Nick at Nite 15 WHDF Florence/Huntsville -CW 648 LATINO TEJANA 174 ABC News Now 179 Fox Business Network 102 NickToons 715 WHDF Florence/Huntsville -CW HD* 644 LATINO TROPICAL 107 Animal Planet 80 Fox College Sports Atlantic 85 Outdoor Channel 25 WHIQ Huntsville - PBS 647 LATINO URBANA 135 BBC America 81 Fox College Sports Central 759 Outdoor Channel HD* 725 WHIQ Huntsville - PBS HD* 608 MAXIMUM PARTY 136 BBC World News 82 Fox College Sports Pacific 242 OWN Oprah Network 19 WHNT Huntsville - CBS 622 NO FENCES 208 BET 156 Fox Movie Channel 106 PBS Kids Sprout 719 WHNT Huntsville - CBS HD* 607 NOTHIN BUT 90’s 213 BET Gospel 178 Fox News Channel 233 Planet Green 20 WHNT Retro TV 641 OPERA PLUS 237 Bravo 76 Fox Soccer Channel 11 QVC 3 WRCB Chattanooga - NBC 602 POP ADULT 103 Cartoon Network 77 Fox Sports South 181 RFD-TV 703 WRCB Chattanooga - NBC HD* 637 POP CLASSICS 687 Cartoon Network HD* 2 FTC Local 138 Science Channel 4 WRCB Retro TV 649 REGIONAL MEXICANA 83 CBS College Sports Ntwk. 203 FUEL TV 69 Shop NBC 45 WTCI Chattanooga - PB S 629 RETRO R&B 212 Centric 124 FX 145 Sleuth 745 WTCI Chattanooga - PBS HD* 646 RETRO LATINO 158 Chiller 147 Game Show Network 225 SOAPnet 9 WTVC Chattanooga - ABC 614 ROCK 200 CMT 202 Great American Country 75 Speed Channel 709 WTVC Chattanooga - ABC HD* 613 ROCK ALTERNATIVE -

Channels You Subscribe to on 71 American Heroes 271 32 ESPNEWS 232 924 Kidz Only! 930 Solid Gold Oldies Your Smartphone, Tablet, Or Laptop
SD Channel HD SD Channel HD ALPHABETICAL CHANNEL GUIDE 116 KCWI3 – Bounce 450 Showtime 550 Watch TV Everywhere 134 KCWI4 – Quest 462 Showtime Showcase 562 11 KDIN – IPTV 211 456 Showtime Extreme 556 SD Channel HD SD Channel HD 110 KDIN2 – IPTV Kids 310 459 Showtime Family 112 KDIN3 – IPTV World 458 Showtime Next 558 FREE With Your CCS Cable TV Package 408 5 Star Max 508 416 Encore Suspense 111 KDIN4 – IPTV Create 454 Showtime SHO x BET 554 929 70’s 420 Encore Westerns 6 KDSM – FOX 206 452 Showtime 2 552 928 80’s EntertainmentStudios.TV HD 365 106 KDSM2 – Comet 460 Showtime Women 560 With Watch TV Everywhere, you can view your favorite programming 927 90’s 30 ESPN 230 107 KDSM3 – Charge 947 Singers & Swing 44 A&E 244 31 ESPN Classic 132 KDSM4 – TBD TV 146 Smile of a Child on your mobile devices wherever you go. Whether you’re waiting 404 Action Max 504 35 ESPN Deportes 39 KFPX – Ion 239 944 Smooth Jazz for a doctor’s appointment, sitting at the airport, or spending your 916 Adult Alternative 33 ESPN U 233 139 KFPX2 – Qubo 2 SMTC TV winter somewhere warmer than Iowa, all you need is an internet 915 Alternative 34 ESPN2 234 140 KFPX3 – ION Life 919 Soft Rock connection and you can access the channels you subscribe to on 71 American Heroes 271 32 ESPNEWS 232 924 Kidz Only! 930 Solid Gold Oldies your smartphone, tablet, or laptop. 25 American Movie Classics 225 154 Eternal World 407 Latino Max 507 143 Son Life Broadcasting 18 Animal Planet 218 484 Flix 28 Lifetime Movie Network 228 941 Sounds of the Seasons Watch live or on demand 70 BBC America