Molecular Diversity Analysis of Tetradium Ruticarpum (Wuzhuyu) In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Umass Extension Landscape Message
Home About Services Landscape Message Weed Herbarium Publications & Resources Education News & Events UMass Extension Landscape Message #18 - Quick Links Scouting Information 2014 Regional Notes Cape Cod Aug 8, 2014 Southeast (Wareham) UMass Extension’s Landscape Message is an educational newsletter intended to inform and guide Green Southeast (Hanson) East Industry professionals in the management of our collective landscape. Scouts compile and record Metro West environmental and phenological data for locations throughout Massachusetts to aid in the monitoring of plant Central and pest development, the planning of management strategies, and the creation of site-specific records for Pioneer Valley future reference. Detailed reports from Extension specialists on growing conditions, pest activity, and cultural Berkshire practices for the management of woody ornamentals, trees, and turf are regular features. UMass Extension has Environmental Data updated the following issue to provide timely management information and the latest regional news and Phenology environmental data. Woody Ornamentals The Landscape Message will be updated bi-weekly July through September. The next message will Landscape Turf be available on August 22. To receive immediate notification when the next Landscape Message update is posted, be sure to join our e-mail list. Archived Messages Scouting Information by Region Regional Notes Cape Cod Region (Barnstable): General Conditions: The Cape missed almost all of the severe weather that hit the rest of the State for this reporting period. The weather has for the most part been sunny and mild, with temperatures averaging in the upper 70s to low 80s F during the day. There were very brief showers on 7/27 that only gave the Cape about 0.07” of rain, barely enough to wet the dust. -

Fragrant Annuals Fragrant Annuals
TheThe AmericanAmerican GARDENERGARDENER® TheThe MagazineMagazine ofof thethe AAmericanmerican HorticulturalHorticultural SocietySociety JanuaryJanuary // FebruaryFebruary 20112011 New Plants for 2011 Unusual Trees with Garden Potential The AHS’s River Farm: A Center of Horticulture Fragrant Annuals Legacies assume many forms hether making estate plans, considering W year-end giving, honoring a loved one or planting a tree, the legacies of tomorrow are created today. Please remember the American Horticultural Society when making your estate and charitable giving plans. Together we can leave a legacy of a greener, healthier, more beautiful America. For more information on including the AHS in your estate planning and charitable giving, or to make a gift to honor or remember a loved one, please contact Courtney Capstack at (703) 768-5700 ext. 127. Making America a Nation of Gardeners, a Land of Gardens contents Volume 90, Number 1 . January / February 2011 FEATURES DEPARTMENTS 5 NOTES FROM RIVER FARM 6 MEMBERS’ FORUM 8 NEWS FROM THE AHS 2011 Seed Exchange catalog online for AHS members, new AHS Travel Study Program destinations, AHS forms partnership with Northeast garden symposium, registration open for 10th annual America in Bloom Contest, 2011 EPCOT International Flower & Garden Festival, Colonial Williamsburg Garden Symposium, TGOA-MGCA garden photography competition opens. 40 GARDEN SOLUTIONS Plant expert Scott Aker offers a holistic approach to solving common problems. 42 HOMEGROWN HARVEST page 28 Easy-to-grow parsley. 44 GARDENER’S NOTEBOOK Enlightened ways to NEW PLANTS FOR 2011 BY JANE BERGER 12 control powdery mildew, Edible, compact, upright, and colorful are the themes of this beating bugs with plant year’s new plant introductions. -

Street Tree Inventory Report Beaumont-Wilshire Neighborhood September 2016 Street Tree Inventory Report: Beaumont-Wilshire Neighborhood September 2016
Street Tree Inventory Report Beaumont-Wilshire Neighborhood September 2016 Street Tree Inventory Report: Beaumont-Wilshire Neighborhood September 2016 Written by: Kat Davidson, Angie DiSalvo, Julie Fukuda, Jim Gersbach, Jeremy Grotbo, and Jeff Ramsey Portland Parks & Recreation Urban Forestry 503-823-4484 [email protected] http://portlandoregon.gov/parks/treeinventory Beaumont-Wilshire Tree Inventory Organizers: Mary Jaron Kelley, Jeff King Staff Neighborhood Coordinator: Jim Gersbach Data Collection Volunteers: Sibylle Baer, Jon Bailey, Ellis Beardsley, Yvonne Boisvert, Rick Burkhard, Dianna Choi, Perry Colclasure, Amanda Cross, Casey Cunningham, Marc Czornic, Greg DiCarlo, Joanne DiCarlo, Monte Edwards, Russell Eng, Jennifer Erickson, Steve Erickson, April Fong, Daniel Goulart, Liz Hay, Sue Hendrick, Starr Hogeboom, Lisa Horowitz, Ben Jones, David Judkins, Dolores Judkins, Mary Jaron Kelley, Jeff King, Ariel Lewin, Chris Lynch, Zosia Lynch, Linda Magnuson, Garth Massey, Sean McMillen, Louis Miles, Mark Miller, Melinda Moeur, Charles Moore, Francis Moore, Majida Nelson, Sheila Nyhus, Laura Paulini, Alexa Pengelly, Maureen Raad, Janine Ragucci, Bruce Richard, Nathan Riggsby, Karen Schaefer, Carl Schwab, Christine Semeniuk, Amy Simpson, Arielle Solomon, Eileen Stark, Jess Tyler, Stephen Upchurch, Matt Vellella, Richard Weber, Ann Wheeler, Paige Witte, Aaron Wolf, and Samantha Wolf Data Entry Volunteers: Rachael Aronson, Max Blasdel, Michael Brehm, Dylan Eglin, Kiel Jenkins, Bethany Lamb, Matthew Pryzborski, Nathan Riggsby, Kristin Sellers, Joshua Sindel, Takayuki Shigematsu, and Eric Watson Arborist-on-Call Volunteers: Ron Davies and Megan Van de Mark GIS Technical Support: Josh Darling, Portland Parks & Recreation Financial Support: Portland Parks & Recreation Cover Photos (from top left to bottom right): 1) Asymmetrical leaves on a hackberry (Celtis occidentalis). 2) The bright pink flowers of a hybrid crape myrtle Lagerstroemia( x indica.) 3) The attractive foliage of a rare Gold Rush dawn redwood (Metasequoia glyptostroboides 'Ogon'). -

Appelhans Et Al Zanthoxylum
Molecular Phylogenetics and Evolution 126 (2018) 31–44 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogeny and biogeography of the pantropical genus Zanthoxylum and its T closest relatives in the proto-Rutaceae group (Rutaceae) ⁎ Marc S. Appelhansa,b, , Niklas Reichelta, Milton Groppoc, Claudia Paetzolda, Jun Wenb a Department of Systematics, Biodiversity and Evolution of Plants, Albrecht-von-Haller Institute of Plant Sciences, University of Goettingen, Untere Karspuele 2, 37073 Goettingen, Germany b Department of Botany, National Museum of Natural History, Smithsonian Institution, P.O. Box 37012, MRC 166, Washington, DC 20013-7012, USA c Departamento de Biologia, Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, São Paulo, Brazil ARTICLE INFO ABSTRACT Keywords: Zanthoxylum L. (prickly ash) is the only genus in the Citrus L. family (Rutaceae) with a pantropical distribution. Bering Land Bridge We present the first detailed phylogenetic and biogeographic study of the genus and its close relatives in the Fagara proto-Rutaceae group. Our phylogenetic analyses based on two plastid and two nuclear markers show that the North Atlantic Land Bridge genus Toddalia Juss. is nested within Zanthoxylum, that earlier generic and intrageneric classifications need Toddalia revision, and that the homochlamydeous flowers of the temperate species of Zanthoxylum are the result of a Transatlantic Disjunction reduction from heterochlamydeous flowers. The biogeographic analyses reveal a Eurasian origin of Zanthoxylum in the Paleocene or Eocene with successive intercontinental or long-range migrations. Zanthoxylum likely crossed the North Atlantic Land Bridges to colonize the Americas in the Eocene, and migrated back to the Old World probably via the Bering Land Bridge in the Oligocene or Miocene. -

Distribution, Biology, Ecology and Control of the Psyllid Diaphorina Citri Kuwayama, a Major Pest of Citrus: a Status Report for China
International Journal of Pest Management, October – December 2006; 52(4): 343 – 352 Distribution, biology, ecology and control of the psyllid Diaphorina citri Kuwayama, a major pest of citrus: A status report for China YUEPING YANG1, MINGDU HUANG1, G. ANDREW C. BEATTIE2, YULU XIA3, GECHENG OUYANG1, & JINJUN XIONG1 1Guangdong Entomological Institute, Guangzhou, Guangdong, People’s Republic of China, 2Centre for Plant and Food Science, University of Western Sydney, Penrith South DC, New South Wales, Australia, and 3National Science Foundation Center for Integrated Pest Management, North Carolina State University, Raleigh, NC, USA Abstract The Asiatic citrus psyllid Diaphorina citri Kuwayama (Hemiptera: Psyllidae) is a major pest of citrus in China. Its status derives, not from the damage it causes, but from its role as the only known vector in China of huanglongbing, a phloem- limited bacterial disease of international importance. The disease can devastate orchards within a few years of planting. It also poses a major threat to endangered indigenous citrus germplasm in Asia and Australasia. The distribution, biology, ecology and control of the psyllid in China are reviewed in these contexts. Constraints and challenges related to control of the vector in China are discussed. Keywords: Diaphorina citri, huanglongbing, distribution, biology, ecology, control gram-negative bacterium Candidatus Liberibacter 1. Introduction asiaticus Jagoueix, Bove´& Garnier (a-Proteobacteria) Asiatic citrus psyllid (Diaphorina citri Kuwayama (Jagoueix et al. 1994; Garnier et al. 2000). ‘Huan- [Hemiptera: Psyllidae]) was recognised as a major glongbing’ is the official name of the disease (van pest of citrus in subtropical and tropical Asia, initially Vuuren 1996) although it has a number of common in India and then elsewhere in the region (Husain and names and is most widely known as citrus greening Nath 1927; Pruthi and Mani 1945; Ebeling 1950). -

Tetradium Danielliidaniellii Bee-Beebee-Bee Tree,Tree, Koreankorean Evodiaevodia
TetradiumTetradium danielliidaniellii Bee-beeBee-bee tree,tree, KoreanKorean EvodiaEvodia Tetradium daniellii(Bee-bee tree, synonym: Euodia daniellii,synonym: Euodia velutina) naturally occurs in China and Korea. The tree was introduced at Kew Gardens, London, England in 1907. It can reach heights of about 20 metres in its original distribution area. With the exception of a few 100-year-old giants, in cultivation this tree will usually stay between 8 - 10 metres tall, with about the same width. The bee-bee tree grows quite quickly and has a wide vase- shaped, semi-open crown on a short stem. In late July, early August, the bee-bee tree is in full bloom with large upright, cream-coloured panicles. For honey bees, wild bees and other insects, the striking, fragrant flowers are very valuable in providing food. After blooming, the tree produces racemes of red-violet capsules. They are easily mistaken for the flowers of the tree, as the colour makes them stand out even more. When the fruits open, they open up to display black seeds. A true delicacy for birds! The compound pinnate leaves make the crown shape transparent, and the tree enchantingly filters light through the crown. The ovoid leaves bud a glossy dark green with a grey-green underside. The leaves turn yellow in autumn. Tetradium daniellii has smooth grey bark (comparable to Fagus sylvatica) and red-brown, downy hairy twigs. Tetradium danielliitolerates acidic and alkaline soils, from sand to clay, and can also handle drought quite well. It is a beautiful tree for future use in parks and large gardens, and - in properly sheltered environments - perfect for use as a characteristic multi-stem tree on a roof garden. -

Bee Friendly: a Planting Guide for European Honeybees and Australian Native Pollinators
Bee Friendly A planting guide for European honeybees and Australian native pollinators by Mark Leech From the backyard to the farm, the time to plant is now! Front and back cover photo: honeybee foraging on zinnia Photo: Kathy Keatley Garvey Bee Friendly A planting guide for European honeybees and Australian native pollinators by Mark Leech i Acacia acuminata © 2012 Rural Industries Research and Development Corporation All rights reserved. ISBN 978 1 74254 369 7 ISSN 1440-6845 Bee Friendly: a planting guide for European honeybees and Australian native pollinators Publication no. 12/014 Project no. PRJ-005179 The information contained in this publication is intended for general use to assist public knowledge and discussion and to help improve the development of sustainable regions. You must not rely on any information contained in this publication without taking specialist advice relevant to your particular circumstances. While reasonable care has been taken in preparing this publication to ensure that information is true and correct, the Commonwealth of Australia gives no assurance as to the accuracy of any information in this publication. The Commonwealth of Australia, the Rural Industries Research and Development Corporation and the authors or contributors expressly disclaim, to the maximum extent permitted by law, all responsibility and liability to any person arising directly or indirectly from any act or omission, or for any consequences of any such act or omission made in reliance on the contents of this publication, whether or not caused by any negligence on the part of the Commonwealth of Australia, RIRDC, the authors or contributors. The Commonwealth of Australia does not necessarily endorse the views in this publication. -

RUTACEAE 芸香科 Yun Xiang Ke Zhang Dianxiang (张奠湘)1; Thomas G
RUTACEAE 芸香科 yun xiang ke Zhang Dianxiang (张奠湘)1; Thomas G. Hartley2, David J. Mabberley3 Shrubs, trees, or sometimes herbs, sometimes scrambling or scandent, sometimes armed, with aromatic volatile oils contained in glands visible at surface of at least leaves, young branchlets, inflorescences, flower parts, fruit, or cotyledons in seed. Stipules absent [or stipular excrescences rarely present]. Leaves alternate, opposite [or whorled], simple (petiole neither apically swollen nor articulate with leaf blade), 1-foliolate (in individual specimens at least some 1-foliolate leaves with petiole apically swollen and/or articulate with leaf blade), or variously compound. Flowers bisexual or unisexual, usually 3–5-merous, actinomorphic or rarely zygomorphic, hypo- gynous [or rarely perigynous]. Perianth in 2 series, with clearly differentiated calyx and corolla or sometimes in 2 irregular series or 1 series, with ± undifferentiated tepals. Sepals distinct or connate to their full length. Petals distinct [or rarely coherent or connate for part of their length]. Stamens usually as many as or 2 × as many as petals or sometimes more numerous; filaments distinct or sometimes coherent or connate for at least part of their length; anthers introrse or sometimes latrorse, longitudinally dehiscent. Disk [rarely lack- ing] within androecium, nectariferous, flattened, annular, cup-shaped, pulvinate, or sometimes columnar, bell-shaped, conic, or hour- glass-shaped. Gynoecium of 1–5 distinct 1-loculed carpels or 2 to many partially to completely connate carpels; placentation axile [very rarely becoming parietal]; ovules 1 to many per locule. Fruit of 2–5 follicles [drupes or samaras] or a single follicle, capsule, or berry [or samara]. Seeds with relatively large embryo; endosperm present and fleshy or lacking. -

Tetradium, De Bijenboom
Binnen de systematische indeling van het regnum vegetabile – het plantenrijk – behoort het geslacht Tetradium tot de orde van de Sapindales en daarbin- nen tot de familie van de Rutaceae, of, in gewoon Nederlands, de ruitachtigen. Andere, wat bekendere geslachten binnen deze grote familie zijn bijvoor- beeld Citrus, Euodia, Phellodendron en Skimmia. Het geslacht Tetradium kent ca. 50 soorten, verspreid over Oost-Azië en hier niet of nauwelijks winterhard, met uitzondering van enkele Chinese exemplaren. Tetradium daniellii is daar één van. Auteur: Jan P. Mauritz VRT Tetradium, de bijenboom Tetradium daniellii is een min of meer struikvor- Het is een prachtige mige, kleine boom van 6 tot maximaal 9 meter hoog in cultuur. Het natuurlijke verspreidings- parkboom met weinig gebied ligt in de Chinese provincies Hubei en Yunnan, tot aan de oostelijke grenzen van de bodemeisen, maar met Himalaya, en in delen van Korea. voorkeur voor een warme, De bijenboom, zoals de Nederlandse naam luidt, vormt een brede, schermvormige kroon met een voedzame grond dikke, vrij korte stam en wijd afstaande gestel- takken. De stam en gesteltakken dragen een vrij middennerf met witte okselbaarden. De herfst- gladde schors, grijs of iets roodachtig van kleur. kleur is schitterend, afhankelijk van de stand- De jonge twijgen zijn vrijwel altijd donzig tot plaats in alle nuances van de kleur geel, variërend dicht, zacht behaard en roodbruin tot bruin van van lichtgoudgeel tot donkeroranjegeel, een heel Jan P. Mauritz, VRT kleur. De beharing verdwijnt geheel, gedeeltelijk bijzondere verschijning, ook dus in de herfst. of vrijwel niet, afhankelijk van de soort. De twij- De bloeiwijze is zeer bijzonder en verschijnt vrijkomen. -
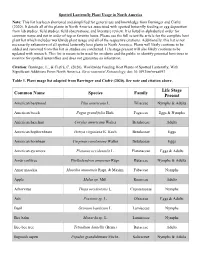
Spotted Lanternfly Plant Usage in North America Note: This List Has Been Shortened and Simplified for General Use and Knowledge from Barringer and Ciafré (2020)
Spotted Lanternfly Plant Usage in North America Note: This list has been shortened and simplified for general use and knowledge from Barringer and Ciafré (2020). It details all of the plants in North America associated with spotted lanternfly feeding or egg deposition from lab studies, field studies, field observations, and literature review. It is listed in alphabetical order for common name and not in order of top or favorite hosts. Please see the full scientific article for the complete host plant list which includes worldwide plant usage and all of the respective citations. Additionally, this list is not necessarily exhaustive of all spotted lanternfly host plants in North America. Plants will likely continue to be added and removed from this list as studies are conducted. Life stage present will also likely continue to be updated with research. This list is meant to be used for residents and the public to identify potential host trees to monitor for spotted lanternflies and does not guarantee an infestation. Citation: Barringer, L., & Ciafré, C. (2020). Worldwide Feeding Host Plants of Spotted Lanternfly, With Significant Additions From North America. Environmental Entomology. doi:10.1093/ee/nvaa093 Table 1. Plant usage list adapted from Barringer and Ciafré (2020). See note and citation above. Life Stage Common Name Species Family Present American basswood Tilia americana L. Tiliaceae Nymphs & Adults American beech Fagus grandifolia Ehrh. Fagaceae Eggs & Nymphs American hazelnut Corylus americana Walter Betulaceae Adults American hophornbeam Ostrya virginiana K. Koch Betulaceae Eggs American hornbean Carpinus caroliniana Walter Betulaceae Eggs American sycamore Platanus occidentalis L. Platanaceae Eggs & Adults Amur corktree Phellodendron amurense Rupr. -

Gardening for Honey Bees by Kathleen M
Gardening for Honey Bees By Kathleen M. Prough Indiana DNR, Div. of Entomology & Plant Pathology You don’t need a large property to start a garden to attract honey bees, or native bees. Take a piece of the lawn that gets 5 to 6 hours of sunlight and turn it into a garden. Think big and start small. Start with a few plants and add more each year. Increase the garden area each year with more plants or propagate the ones you have. If you run out of garden space start giving plants or seeds to neighbors. When starting the garden strive for several things: Abundance - Clump plants so the bees can find and visit many flowers in one location. This provides a worthwhile food stop! Coneflowers in a large clump will attract honey bees and all types of native bees and butterflies. Sequence – Plant for bloom succession. You need a succession of overlapping blooming from spring to fall. Learn when specific plants bloom so you get a good overlap. Diversity - Select plants that will provide pollen as well as nectar. Bees need diverse pollen and nectar sources for balanced diet. Don’t forget trees and shrubs that can provide pollen and nectar. Pesticide use - If possible, do not use pesticides! Insecticides can kill beneficial insects as well as the insect you are trying to kill. Herbicides may kill plants (dandelions and Dutch white clover) that bees can get pollen and nectar from. So, do not go for that perfect lawn with no weeds in it. Plant traits that may attract bees: Flower color- bright white, yellow, blue or UV Flower shape- shallow, have a landing platform, tubular, single flower top Nectar guides present- guides the bees into the plant Nectar is present- usually fresh, mild and a pleasant smell Pollen- often sticky and scented Observe the bees closely to learn the plants from which they collect nectar and pollen.