Meiotic Recombination: the Essence of Heredity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogenetic Investigations of Sordariaceae Based on Multiple Gene Sequences and Morphology
mycological research 110 (2006) 137– 150 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/mycres Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology Lei CAI*, Rajesh JEEWON, Kevin D. HYDE Centre for Research in Fungal Diversity, Department of Ecology & Biodiversity, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, PR China article info abstract Article history: The family Sordariaceae incorporates a number of fungi that are excellent model organisms Received 10 May 2005 for various biological, biochemical, ecological, genetic and evolutionary studies. To deter- Received in revised form mine the evolutionary relationships within this group and their respective phylogenetic 19 August 2005 placements, multiple-gene sequences (partial nuclear 28S ribosomal DNA, nuclear ITS ribo- Accepted 29 September 2005 somal DNA and partial nuclear b-tubulin) were analysed using maximum parsimony and Corresponding Editor: H. Thorsten Bayesian analyses. Analyses of different gene datasets were performed individually and Lumbsch then combined to generate phylogenies. We report that Sordariaceae, with the exclusion Apodus and Diplogelasinospora, is a monophyletic group. Apodus and Diplogelasinospora are Keywords: related to Lasiosphaeriaceae. Multiple gene analyses suggest that the spore sheath is not Ascomycota a phylogenetically significant character to segregate Asordaria from Sordaria. Smooth- Gelasinospora spored Sordaria species (including so-called Asordaria species) constitute a natural group. Neurospora Asordaria is therefore congeneric with Sordaria. Anixiella species nested among Gelasinospora Sordaria species, providing further evidence that non-ostiolate ascomata have evolved from ostio- late ascomata on several independent occasions. This study agrees with previous studies that show heterothallic Neurospora species to be monophyletic, but that homothallic ones may have a multiple origins. -

Meiosis As an Evolutionary Adaptation for DNA Repair
19 Meiosis as an Evolutionary Adaptation for DNA Repair Harris Bernstein1, Carol Bernstein1 and Richard E. Michod2 1Department of Cellular and Molecular Medicine, University of Arizona 2Department of Ecology and Evolutionary Biology, University of Arizona USA 1. Introduction The adaptive function of sex remains, today, one of the major unsolved problems in biology. Fundamental to achieving a resolution of this problem is gaining an understanding of the function of meiosis. The sexual cycle in eukaryotes has two key stages, meiosis and syngamy. In meiosis, typically a diploid cell gives rise to haploid cells. In syngamy (fertilization), typically two haploid gametes from different individuals fuse to generate a new diploid individual. A unique feature of meiosis, compared to mitosis, is recombination between non-sister homologous chromosomes. Usually these homologous chromosomes are derived from different individuals. In mitosis, recombination can occur, but it is ordinarily between sister homologs, the two products of a round of chromosome replication. Birdsell & Wills (2003) have reviewed the various hypotheses for the origin and maintenance of sex and meiotic recombination, including the hypothesis that sex is an adaptation for the repair of DNA damage and the masking of deleterious recessive alleles. Recently, we presented evidence that among microbial pathogens, sexual processes promote repair of DNA damage, especially when challenged by the oxidative defenses of their biologic hosts (Michod et al., 2008). Here, we present evidence that meiosis is primarily an evolutionary adaptation for DNA repair. Since our previous review of this topic (Bernstein et al., 1988), there has been a considerable increase in relevant information at the molecular level on the DNA repair functions of meiotic recombination, and this new information is emphasized in the present chapter. -

MEIOSIS and RECOMBINATION in SORDARIA FIMICOLA Introduction
MEIOSIS AND RECOMBINATION IN SORDARIA FIMICOLA Introduction: In ascomycete fungi, a form of meiosis occurs in which the products of meiosis order themselves within a fruiting body according to the physical separation and segregation of chromatids during the meiotic process. This is covered in some detail on pages 150-152 (including Figures 4.26 and 4.27) in Hartl and Jones, Essential Genetics. You should study these pages before beginning this module. As described, ordered tetrad analysis provides a way to measure the genetic map distance between a gene and the centromere of the chromosome on which that gene resides. That is what you will do over the next two weeks in this laboratory. I. Natural history and Life Cycle of Sordaria fimicola Sordaria fimicola is an ascomycete fungi that can be found growing in rotting vegetation and animal dung (in fact, the name Sordaria fimicola means "filthy dung dweller"). Sordaria and another ascomycete, the common bread fungus Neurospora crassa (Fig. 4.26), have been used as model systems for studying the process of chromosome exchange (crossing-over) because of their reproductive characteristics. The life cycle of Sordaria is representative of the ascomycetes (although there are substantial differences in the details among species). The individual fungus begins as a haploid ascospore. The ascospore germinates to form hyphae (singular = hypha), which are long filaments comprised of haploid cells. These hyphae grow and extend throughout the nutrient source (dung or rotting vegetation in nature, nutrient medium in the laboratory situation) and digest it by means of enzymes secreted by the cells. Nutrients are then absorbed into the cells. -

Coprophilous Fungal Community of Wild Rabbit in a Park of a Hospital (Chile): a Taxonomic Approach
Boletín Micológico Vol. 21 : 1 - 17 2006 COPROPHILOUS FUNGAL COMMUNITY OF WILD RABBIT IN A PARK OF A HOSPITAL (CHILE): A TAXONOMIC APPROACH (Comunidades fúngicas coprófilas de conejos silvestres en un parque de un Hospital (Chile): un enfoque taxonómico) Eduardo Piontelli, L, Rodrigo Cruz, C & M. Alicia Toro .S.M. Universidad de Valparaíso, Escuela de Medicina Cátedra de micología, Casilla 92 V Valparaíso, Chile. e-mail <eduardo.piontelli@ uv.cl > Key words: Coprophilous microfungi,wild rabbit, hospital zone, Chile. Palabras clave: Microhongos coprófilos, conejos silvestres, zona de hospital, Chile ABSTRACT RESUMEN During year 2005-through 2006 a study on copro- Durante los años 2005-2006 se efectuó un estudio philous fungal communities present in wild rabbit dung de las comunidades fúngicas coprófilos en excementos de was carried out in the park of a regional hospital (V conejos silvestres en un parque de un hospital regional Region, Chile), 21 samples in seven months under two (V Región, Chile), colectándose 21 muestras en 7 meses seasonable periods (cold and warm) being collected. en 2 períodos estacionales (fríos y cálidos). Un total de Sixty species and 44 genera as a total were recorded in 60 especies y 44 géneros fueron detectados en el período the sampling period, 46 species in warm periods and 39 de muestreo, 46 especies en los períodos cálidos y 39 en in the cold ones. Major groups were arranged as follows: los fríos. La distribución de los grandes grupos fue: Zygomycota (11,6 %), Ascomycota (50 %), associated Zygomycota(11,6 %), Ascomycota (50 %), géneros mitos- mitosporic genera (36,8 %) and Basidiomycota (1,6 %). -

The Pingry Community Research (PCR) Journal
The Pingry Community Research (PCR) Journal A Journal of Scientific Research at The Pingry School Volume 2: Spring 2014 Page 1: Siemens and Intel WinnersContents By Akash Kumar’17 2: How Naked Mole Rats May Help the War on Cancer By Brad Hong’16 3: The Role of Alcohol Dosage on HPA Axis Functionality By Jackson Artis’16, Julia Friend’15, Avery Hatfield’14, and Luke De 5: The Effects of Sleep Deprivation on Anxiety in Danio rerio By Stacy Chen’14, Kathleen Murray’15, and Allison Yu’14 7: Soil Variations to Maximize Arabidopsis thaliana Yield By Erica Cheung’14, Tammy Gu’14, Rabia Khan’14 9: Melanin Protects Sordaria fimicola Spores From Ultraviolet Light By Katherine Curran’14 11: The Effects of Various Iron Concentrations on Algal Growth By Rachel Davis’14 and Melanie Naratil’14 13: Recycling Waste CO2 to Grow Algae By Sofia Deak’14, Alli Dorneo’14, and Ben Kaminoff’14 15: The Effect of UV Radiation on Bacterial Tolerance in Escherichia coli By Natalie Gilbert’14, Stephanie Yeh’14, and Aigner Mizzelle’14 16: Are We as Smart as We Think We Are? By Lauren Graves’14 18: CG-3634 Notch Phenotype Screen By Amol Kapoor’14 21: Purification of Salmonella Transcription Factors HilD and HilC By Teddy Leithead’14, Elizabeth Kraeutler’15, Jessica Day, F. John Kull, and Morgan D’Ausilio 22: Modeling ToxT to Explain How Cholera Toxicity can be Regulated by Fatty Acids: The 2014 Pingry SMART Team Project By Rachel Wu’16 and Emily Kwon’16 25: Effect of Methane Digesters on Global Greenhouse Gases By Pradyuth Maganti’15 and Matthew Rice’15 27: Effect of Coffee Grounds on Lettuce Growth By Rebecca Muller’14 and Lauren Ru’14 29: Comparing the Effects of Simple and Complex Fish Diets on Lettuce (Lactuca sativa) Growth By Christina Ou’15 Page 30: Using Olive Oil to Improve the Effectiveness of Nepetalactone as an Insect Repellant By Adriano Taglietti’14 and Charlie Wollmuth’14 32: Protein Tyrosine Phosphatase S (PTPRS) Is a Growth Suppressor in Lung Adenocarcinoma Cell Lines By L. -

Genetic Recombination at the Buff Spore Color Locus in Sordaria Brevicollis
Copyright 0 1983 by the Genetics Society of America GENETIC RECOMBINATION AT THE BUFF SPORE COLOR LOCUS IN SORDARIA BREVICOLLIS. 11. ANALYSIS OF FLANKING MARKER BEHAVIOR IN CROSSES BETWEEN BUFF MUTANTS HELEN SANG' AND HAROLD L. K. WHITEHOUSE Botany School, University of Combridge, Cambridge, England Manuscript received February 24, 1982 Revised copy accepted July 21, 1982 ABSTRACT Aberrant asci containing one or more wild-type spores were selected from crosses between pairs of alleles of the buff locus in the presence of closely linked flanking markers. Data were obtained relating to the site of aberrant segregation and the position of any associated crossover giving recombination of flanking markers. Aberrant segregation at a proximal site within the buff gene may be associated with a crossover proximal to the site of aberrant segregation or, with equal frequency, with a crossover distal to the site of the second mutant present in the cross. Similarly, segregation at a distal site may be associated with a crossover distal to the site or, with lower frequency, with a crossover proximal to the site of the proximal mutant present in the cross. Crossovers between the alleles were rare. This evidence for the relationship between hybrid DNA and crossing over is discussed in terms of current models for the mechanism of recombination. URRENT models of the mechanism of genetic recombination consider gene C conversion and crossing over to be consequences of the same primary event. The MESELSONand RADDING(1975) model proposes that hybrid DNA formation is initiated asymmetrically outside a gene by single-strand transfer between paired homologues. -

Savoryellales (Hypocreomycetidae, Sordariomycetes): a Novel Lineage
Mycologia, 103(6), 2011, pp. 1351–1371. DOI: 10.3852/11-102 # 2011 by The Mycological Society of America, Lawrence, KS 66044-8897 Savoryellales (Hypocreomycetidae, Sordariomycetes): a novel lineage of aquatic ascomycetes inferred from multiple-gene phylogenies of the genera Ascotaiwania, Ascothailandia, and Savoryella Nattawut Boonyuen1 Canalisporium) formed a new lineage that has Mycology Laboratory (BMYC), Bioresources Technology invaded both marine and freshwater habitats, indi- Unit (BTU), National Center for Genetic Engineering cating that these genera share a common ancestor and Biotechnology (BIOTEC), 113 Thailand Science and are closely related. Because they show no clear Park, Phaholyothin Road, Khlong 1, Khlong Luang, Pathumthani 12120, Thailand, and Department of relationship with any named order we erect a new Plant Pathology, Faculty of Agriculture, Kasetsart order Savoryellales in the subclass Hypocreomyceti- University, 50 Phaholyothin Road, Chatuchak, dae, Sordariomycetes. The genera Savoryella and Bangkok 10900, Thailand Ascothailandia are monophyletic, while the position Charuwan Chuaseeharonnachai of Ascotaiwania is unresolved. All three genera are Satinee Suetrong phylogenetically related and form a distinct clade Veera Sri-indrasutdhi similar to the unclassified group of marine ascomy- Somsak Sivichai cetes comprising the genera Swampomyces, Torpedos- E.B. Gareth Jones pora and Juncigera (TBM clade: Torpedospora/Bertia/ Mycology Laboratory (BMYC), Bioresources Technology Melanospora) in the Hypocreomycetidae incertae -

Demonstration of Crossing-Over During Meiosis in Sordaria Fimicola
1 Demonstration of crossing-over during meiosis in Sordaria fimicola Introduction Sexual reproduction is a special case of cell reproduction in which the genes of two individuals are “shuffled.” In this way it is unlike asexual reproduction, in which all the daughter cells (and organisms) are genetically identical to the parent cell. The result of asexual reproduction is a series of clones (rather like the bacterial colonies we observed last week). In contrast, sexual reproduction involves the fusion of two cells called gametes, one typically understood as male, the other as female (sperm and eggs in humans). Gametes have half the typical number of chromosomes. For example, if the species typically has 46 chromosomes, like humans, then the gametes will have 23. The reduction in the number of chromosomes occurs during a specialized cell division called meiosis. The choice of which chromosomes (those originally inherited from the father or from the mother, paternal or maternal, respectively) are found in which gamete is a random event. Thus depending on N, the total number of kinds of chromosomes in the genome, there are 2N different combinations of paternal and maternal chromosomes that can arise due solely to the divisions of meiosis. Additional variation arises from two other sources: mutation and recombination. We have already had some (disappointing) evidence of the rarity of mutation (i.e. our failure to detect any ampicillin-resistant mutations in a total of 0.3 mL of a population containing about 3 x 109 E. coli cells per mL.) Today we will discover a system that allows us to estimate recombination, and indirectly a means of mapping the chromosome. -

Fungal Recombination TERRY L
MICROBIOLOGICAL REVIEWS, Mar. 1985, p. 33-58 Vol. 49, No. 1 0146-0749/85/010033-26$02.00/0 Copyright © 1985, American Society for Microbiology Fungal Recombination TERRY L. ORR-WEAVERt* AND JACK W. SZOSTAKt Dana-Farber Cancer Institute and Department ofBiological Chemistry, Harvard Medical School, Boston, Massachusetts 02115 INTRODUCTION .33.......................3 MEIOTIC RECOMBINATION .33 Gene Conversion and Postmeiotic Segregation .33 Heteroduplex DNA and Meiotic Recombination .34 Co-Conversion .......... 35 Allele-Specific Segregation Patterns .36 Polarity .38 Crossing Over and Aberrant Segregation .39 Recombination Initiation Sites .40 Mutations Affecting Meiotic Recombination.41 Role and Timing of Recombination in Meiosis .42 MITOTIC RECOMBINATION .43 Induction of Mitotic Recombination .43 Recombination at the Two-Strand Stage .43 Symmetric Heteroduplex DNA.44 Length of Heteroduplex DNA Tracts .44 Association of Crossing Over.46 Recombination Mutants .46 Comnparison of Meiotic and Mitotic Recombination .46 SPECIFIC RECOMBINATION EVENTS .47 Plasmid-Chromosome Recombination.47 Mating-Type Switching .48 2,Im Recombination .48 ENZYMOLOGY OF RECOMBINATION .49 RECOMBINATION MODELS .49 49 The Holliday Model ............................... The Meselson-Radding Model .............................. 4 50 The Double-Strand Break Repair Model ............................................................. 51 Comparison of Meselson-Radding and Double-Strand Break Repair Models.52 CONCLUDING REMARKS .54 ACKNOWLEDGMENTS .55 LITERATURE CITED .55 INTRODUCTION in meiosis and in vegetative growth and contrast the nature Despite many years of intensive genetic analysis, the of meiotic and mitotic recombination. We review plasmid- molecular mechanisms of the recombination events that chromosome recombination and, several site-specific recomi- have been studied in the fungi remain unknown. Numerous bination events in yeast cells and describe attempts to defihe models have been proposed to explain the genetic data, and recombination in enzymological terms. -

Diversity and Distribution Patterns of Endolichenic Fungi in Jeju Island, South Korea
sustainability Article Diversity and Distribution Patterns of Endolichenic Fungi in Jeju Island, South Korea Seung-Yoon Oh 1,2 , Ji Ho Yang 1, Jung-Jae Woo 1,3, Soon-Ok Oh 3 and Jae-Seoun Hur 1,* 1 Korean Lichen Research Institute, Sunchon National University, 255 Jungang-Ro, Suncheon 57922, Korea; [email protected] (S.-Y.O.); [email protected] (J.H.Y.); [email protected] (J.-J.W.) 2 Department of Biology and Chemistry, Changwon National University, 20 Changwondaehak-ro, Changwon 51140, Korea 3 Division of Forest Biodiversity, Korea National Arboretum, 415 Gwangneungsumok-ro, Pocheon 11186, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-61-750-3383 Received: 24 March 2020; Accepted: 1 May 2020; Published: 6 May 2020 Abstract: Lichens are symbiotic organisms containing diverse microorganisms. Endolichenic fungi (ELF) are one of the inhabitants living in lichen thalli, and have potential ecological and industrial applications due to their various secondary metabolites. As the function of endophytic fungi on the plant ecology and ecosystem sustainability, ELF may have an influence on the lichen diversity and the ecosystem, functioning similarly to the influence of endophytic fungi on plant ecology and ecosystem sustainability, which suggests the importance of understanding the diversity and community pattern of ELF. In this study, we investigated the diversity and the factors influencing the community structure of ELF in Jeju Island, South Korea by analyzing 619 fungal isolates from 79 lichen samples in Jeju Island. A total of 112 ELF species was identified and the most common species belonged to Xylariales in Sordariomycetes. -

Emarcea Castanopsidicola Gen. Et Sp. Nov. from Thailand, a New Xylariaceous Taxon Based on Morphology and DNA Sequences
STUDIES IN MYCOLOGY 50: 253–260. 2004. Emarcea castanopsidicola gen. et sp. nov. from Thailand, a new xylariaceous taxon based on morphology and DNA sequences Lam. M. Duong2,3, Saisamorn Lumyong3, Kevin D. Hyde1,2 and Rajesh Jeewon1* 1Centre for Research in Fungal Diversity, Department of Ecology & Biodiversity, The University of Hong Kong, Pokfulam Road, Hong Kong, SAR China; 2Mushroom Research Centre, 128 Mo3 Ban Phadeng, PaPae, Maetaeng, Chiang Mai 50150, Thailand 3Department of Biology, Chiang Mai University, Chiang Mai, Thailand *Correspondence: Rajesh Jeewon, [email protected] Abstract: We describe a unique ascomycete genus occurring on leaf litter of Castanopsis diversifolia from monsoonal forests of northern Thailand. Emarcea castanopsidicola gen. et sp. nov. is typical of Xylariales as ascomata develop beneath a blackened clypeus, ostioles are papillate and asci are unitunicate with a J+ subapical ring. The ascospores in Emarcea cas- tanopsidicola are, however, 1-septate, hyaline and long fusiform, which distinguishes it from other genera in the Xylariaceae. In order to substantiate these morphological findings, we analysed three sets of sequence data generated from ribosomal DNA gene (18S, 28S and ITS) under different optimality criteria. We analysed this data to provide further information on the phylogeny and taxonomic position of this new taxon. All phylogenies were essentially similar and there is conclusive mo- lecular evidence to support the establishment of Emarcea castanopsidicola within the Xylariales. Results indicate that this taxon bears close phylogenetic affinities to Muscodor (anamorphic Xylariaceae) and Xylaria species and therefore this genus is best accommodated in the Xylariaceae. Taxonomic novelties: Emarcea Duong, R. Jeewon & K.D. -
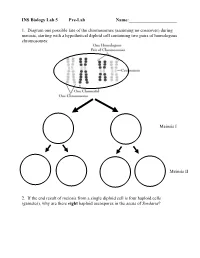
INS Biology Lab 5 Pre-Lab Name:______
INS Biology Lab 5 Pre-Lab Name:_____________________ 1. Diagram one possible fate of the chromosomes (assuming no crossover) during meiosis, starting with a hypothetical diploid cell containing two pairs of homologous chromosomes: Meiosis I Meiosis II 2. If the end result of meiosis from a single diploid cell is four haploid cells (gametes), why are there eight haploid ascospores in the ascus of Sordaria? 2 Winter Lab 1. Meiosis This lab exercise courtesy of Mike Clayton, University of Wisconsin Adapted from a lab written by Jon Glase, Cornell University I. The Process of Meiosis Meiosis is a process of two sequenced nuclear divisions neither of which is identical to mitosis. Meiosis always begins with a nucleus with two sets of chromosomes (a diploid nucleus) where each chromosome has two chromatids. Meiosis produces four nuclei each with one set of chromosomes (haploid nuclei) in which each chromosome has one chromatid. Meiosis together with syngamy constitutes sexuality in eukaryotes. Sexual reproduction generates individuals with new and unique genotypic combinations each generation. Meiosis, by itself, can generate new combinations of genetic material. It does so in two ways. -- through the independent assortment of different homologous pairs of chromosomes and -- through crossing over between homologous pairs of chromosomes. Pop Beads as Models of Chromosomes The shuffling and recombination of genetic material can be modeled using pop beads. Before lab, preview this web page to see a demonstration of meiosis using pop beads ( http://botit.botany.wisc.edu/courses/botany_130/Meiosis/pop.html ). In lab, you will work with a partner to recreate the following scenarios of meiosis..