Exploring and Developing Algorithm of Predicting Advanced Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CDIBC 11 Digestivehepato SG Download Resource
Key Diseases Associated with the Digestive and Hepatobiliary Systems Study Guide 1.1 Key Diseases Associated with the Digestive, Hepatobiliary and Urinary Systems © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.3 Human Digestive System 1.4 Documentation for GI Conditions (cont.) © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.5 Abdominal Pain 1.6 Conditions Associated W/Abdominal Pain © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.7 MS-DRG 391, 392 1.8 Abdominal Pain: Epigastric © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.9 Abdominal Pain: Epigastric (cont.) 1.10 Abdominal Pain: Gastritis © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.11 Gastritis/Duodenitis/Gastroduodenitis 1.12 Gastritis © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.13 Ulcerative Colitis 1.14 Ulcerative Colitis: K51 © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.15 Crohn’s Disease 1.16 Coding Clinic, 4th Quarter 2012 © 2017 HCPro. All rights reserved. These materials may not be duplicated without the express written permission of HCPro. 1.17 Coding Clinic, 4th Quarter 2012 (cont.) 1.18 Ulcer of the Intestine © 2017 HCPro. All rights reserved. -

Human Body- Digestive System
Previous reading: Human Body Digestive System (Organs, Location and Function) Science, Class-7th, Rishi Valley School Next reading: Cardiovascular system Content Slide #s 1) Overview of human digestive system................................... 3-4 2) Organs of human digestive system....................................... 5-7 3) Mouth, Pharynx and Esophagus.......................................... 10-14 4) Movement of food ................................................................ 15-17 5) The Stomach.......................................................................... 19-21 6) The Small Intestine ............................................................... 22-23 7) The Large Intestine ............................................................... 24-25 8) The Gut Flora ........................................................................ 27 9) Summary of Digestive System............................................... 28 10) Common Digestive Disorders ............................................... 31-34 How to go about this module 1) Have your note book with you. You will be required to guess or answer many questions. Explain your guess with reasoning. You are required to show the work when you return to RV. 2) Move sequentially from 1st slide to last slide. Do it at your pace. 3) Many slides would ask you to sketch the figures. – Draw them neatly in a fresh, unruled page. – Put the title of the page as the slide title. – Read the entire slide and try to understand. – Copy the green shade portions in the note book. 4) -

Digestive System (Part-I)
NPTEL – Basic Courses – Basic Biology Lecture 11: Digestive System (Part-I) Introduction: The collective processes by which a living organism takes food which are necessary for their growth, maintenance and energy needs is called nutrition. The chemical substances present in the food are called nutrients. DIFFERENT MODE OF NUTRITION IN ORGANISMS: It is important to know the different modes of nutrition in all living organisms in order to understand energy flow within the ecosystem. Plant produces high energy organic food from inorganic raw materials. They are called autotroph and the mode of nutrition is known as autotrophic nutrition. Animals feed on those high energy organic food, are called as heterotrophs and their mode of nutrition is known as heterotrophic nutrition Heterotrophic nutrition further sub-categorise in holozoic, parasitic, and saprophytic mode of nutrition based on the pattern and class of food that is taken inside. Holozoic Nutrition: It involves taking entire organic food and this can be in the form of whole part of plant or animal. Most of the free living protozoans, humans and other animals fall under this category. Saprophytic Nutrition: The organism fulfils the requirement of food from the rotten parts of dead organisms and decaying matter. The organisms secrete digestive enzymes outside the body on their food and then take in digested food. It is a kind of extra-cellular digestion. Examples: Housefly, Spiders etc. Parasitic Nutrition: The organism fulfils the requirement of food from the body of another organism. The parasites are of two distinct types, one which lives inside the host and the other which lives outside. -

Potential Impacts of Gut Microbiota on Immune System Related Diseases: Current Studies and Future Challenges
Acta Medica 2018; 49(2): 31-37 acta medica REVIEW Potential Impacts of Gut Microbiota on Immune System Related Diseases: Current Studies and Future Challenges 1 Tayfun Hilmi Akbaba , [BSc] ABSTRACT Banu Balcı Peynircioğlu *, [MD] Human microbiota includes trillions of cohabitant microorganisms in a symbiotic re- lationship. They directly or indirectly communicate with immune system. Human microbiome profiling studies has accelerated microbiota studies and interest to mi- 1, Hacettepe University, Faculty of Medicine, crobiota and disease relationship . Some metabolic activities of human microbiota Department of Medical Biology, Ankara, Turkey were known, but in recent years many different roles in addition to metabolic activi- ties, have been shown. It can affect systems and mechanisms, and most importantly clinical course of diseases in dysbiosis condition, and reduces most symptoms when * Corresponding Author: Banu Balci- Peynircioğlu. symbiosis is provided. These features make microbiota a potential therapeutic tool or [MD] Hacettepe University Faculty of Medicine, biomarker for a spate of disease in clinic applications.This review summarizes host- Department of Medical Biology, Ankara – Turkey gut microbiota interaction, role of microbiota in immune-related diseases, and poten- e-mail: [email protected] tial therapeutic approaches. Phone: +903122102541 Keywords: Gut microbiota, microbiome, immune system related disease Received: 11 April 2018, Accepted: 29 May 2018, Published online: 29 June 2018 INTRODUCTION The human microbiota consists of bacteria, archaea, of highly conserved sequence, and taxonomic pro- viruses and eukaryotic microbes that live in and on files, whole genome shogun (WGS) or metagenom- our bodies, and corporate genome of our micro- ics sequencing of whole community DNA, which bial symbionts, commensal and pathobiont (po- make easy to study microbiome. -

Carcinoembryonic Antigen in Serum in Diseases of the Liver and Pancreas
J Clin Pathol: first published as 10.1136/jcp.26.7.470 on 1 July 1973. Downloaded from J. clin. Path., 1973, 26, 470-475 Carcinoembryonic antigen in serum in diseases of the liver and pancreas S. K. KHOO AND I. R. MACKAY From the Clinical Research Unit of The Walter and Eliza Hall Institute of Medical Research and The Royal Melbourne Hospital, Victoria 3050, Australia SYNOPSIS Carcinoembryonic antigen (CEA) was measured in whole serum and in serum extracted with perchloric acid by microradioimmunoassay in patients with benign and malignant diseases of the liver and pancreas. The level of detectability was 5 ng per ml. This level or greater was present in the serum of 50 % of patients with chronic diffuse liver disease, 640% with pancreatitis, 94o/% with cancer of the digestive system, and 3% ofcontrols. The incidence of levels of CEA of 5 ng/ml or more differed for various categories of chronic liver disease: from 22 % in active chronic hepatitis, 46 % in primary biliary cirrhosis, 63 % in hepatoma, 78 % in cryptogenic cirrhosis, and 88 % in alcoholic cirrhosis; levels of CEA correlated with degrees of impairment of liver function as judged by bromsulphalein retention and serum levels of alkaline phosphatase and transaminase. In pancreatitis, 64% of cases had levels of CEA ranging from 5 to 20 ng/ml and in cancer of the pancreas 94 % had levels above 5 ng/ml and 50 % above 20 ng/ml. copyright. Carcinoembryonic antigen (CEA) in serum was CEA in patients with colonic cancer from 1904, originally held to be specific for cancer of the through 5300 to 100%; equivalent findings were digestive system (Gold and Freedman, 1965a, reported in a joint investigation (1972) on CEA in 1965b), but CEA-like activity has been reported in colonic cancer. -

Circulating Carcinoembryonic Antigen (CEA): Inflammatory Bowel Disease
Gut: first published as 10.1136/gut.14.11.880 on 1 November 1973. Downloaded from Gut, 1973, 14, 880-884 Circulating carcinoembryonic antigen (CEA): Relationship to clinical status of patients with inflammatory bowel disease A. H. RULE, C. GOLESKI-REILLY, D. B. SACHAR1, J. VANDEVOORDE, AND H. D. JANOWITZ From the Division of Gastroenterology, Department ofMedicine, The Mount Sinai Hospital and The Mount Sinai School ofMedicine of the City Untiversity ofNew York, New York, USA SUMMARY Plasma levels of circulating carcinoembryonic antigen (CEA) were measured by zirconyl phosphate gel radioimmunoassay in 112 patients with chronic inflammatory bowel disease. The levels were then related to category, extent, duration, and severity of disease, as well as to the ages and surgical status of the patients. The distribution of CEA levels and their mean values were significantly raised over the levels in 33 normal control subjects, and were similar among patients with ulcerative colitis compared with those with granulomatous bowel disease. Positive values were defined as those in excess of 2.5 ng/ml. Positive assays occurred in 42 % of ulcerative colitis patients, in 38 % of Crohn's disease patients, and in 40 % of the total group with inflammatory bowel disease. Among normal control subjects, only 3 % were positive. Among inflammatory bowel disease patients, positive CEA assays occurred more frequently with more severe disease, more extensive anatomical involvement, younger ages, and shorter duration of disease. Those patients who had undergone http://gut.bmj.com/ total colectomy showed levels of circulating CEA and frequency of CEA positivity similar to those of an age-matched normal control group. -

A Giving Smarter Guide
0 AUTHORS LEAD AUTHOR Erik Lontok, PhD CONTRIBUTING AUTHORS LaTese Briggs, PhD Sonya B. Dumanis, PhD YooRi Kim, MS Ekemini A.U. Riley, PhD Danielle Salka Melissa Stevens, MBA IBD SCIENTIFIC ADVISORY GROUP We graciously thank the members of the IBD Scientific Advisory Group for their participation and contribution to the IBD Project and Giving Smarter Guide. The informative discussions before, during, and after the IBD retreat were critical to identifying the unmet needs and philanthropic opportunities to benefit patients and advance IBD research. Mark Allegretta, PhD Michael Fischbach, PhD Associate Vice President, Commercial Research Associate Professor, Division of Bioengineering National Multiple Sclerosis Society and Therapeutic Sciences University of California, San Francisco Eugene B. Chang, MD Martin Boyer Professor of Medicine Kevin Grimes, MD Department of Medicine Associate Professor, Chemical and Systems Knapp Center for Biomedical Discovery Biology; Co-Director, SPARK Program University of Chicago Stanford School of Medicine Jean-Frederic Colombel, MD Stephen B. Hanauer, MD Director, The Leona M. and Harry B. Helmsley Medical Director, Digestive Health Clinic Charitable Trust IBD Center Professor of Medicine, Gastroenterology and Professor of Medicine, Gastroenterology Hepatology Mount Sinai Hospital Northwestern University Feinberg School of Medicine Tejal A. Desai, PhD Professor and Chair, Department of Caren Heller, MD, MBA Bioengineering and Therapeutic Sciences Chief Scientific Officer University of California, San Francisco -

Mammalian Gastrointestinal Tract Parameters Modulating the Integrity, Surface Properties, and Absorption of Food-Relevant Nanoma
Advanced Review Mammalian gastrointestinal tract parameters modulating the integrity, surface properties, and absorption of food-relevant nanomaterials Susann Bellmann,1 David Carlander,2 Alessio Fasano,3 Dragan Momcilovic,4 Joseph A. Scimeca,5 W. James Waldman,6 Lourdes Gombau,7 Lyubov Tsytsikova,8 Richard Canady,8 Dora I. A. Pereira9 and David E. Lefebvre10∗ Many natural chemicals in food are in the nanometer size range, and the selective uptake of nutrients with nanoscale dimensions by the gastrointestinal (GI) tract is a normal physiological process. Novel engineered nanomaterials (NMs) can bring various benefits to food, e.g., enhancing nutrition. Assessing potential risks requires an understanding of the stability of these entities in the GI lumen, and an understanding of whether or not they can be absorbed and thus become systemically available. Data are emerging on the mammalian in vivo absorption of engineered NMs composed of chemicals with a range of properties, including metal, mineral, biochemical macromolecules, and lipid-based entities. In vitro and in silico fluid incubation data has also provided some evidence of changes in particle stability, aggregation, and surface properties following interaction with luminal factors present in the GI tract. The variables include physical forces, osmotic concentration, pH, digestive enzymes, other food, and endogenous biochemicals, and commensal microbes. Further research is required to fill remaining data gaps on the effects of these parameters on NM integrity, physicochemical properties, and GI absorption. Knowledge of the most influential luminal parameters will be essential when developing models of the GI tract to quantify the percent absorption of food-relevant engineered NMs for risk assessment. -

All About the GI System
All about The GI System A self paced learning program for direct care staff By: Barbara Obst, RN, MS Fall 2011 Introduction • This is a self paced independent training program for direct care staff. • The information taught will promote safe care of individuals served in the DD community setting. Objectives:The Participant will be able to: • Understand the basic anatomy and physiology of the gastrointestinal tract. • Identify two types of constipation. • State 4 causes of constipation. • State 3 causes of diarrhea. • Indentify the reason why the Bristol Stool Scale is important. • Identify 3 possible solutions to prevent constipation. The GI System a Snapshot The Human Digestive System • A complex system of organs and glands that work together to digest (breakdown) food into smaller particles that are absorbed to support the body’s nutritional needs. • The digestive system concentrates the waste/by-products of the digestive process and expels the waste products in the form of feces/stool. • Most of the digestive system is a tube-like tract that propels the food through the body. The Digestive System • The digestive system is a tube-like tract that includes the esophagus, stomach and lpch.org intestines. This tract runs from the mouth to the anus. • In addition to the tube-like tract, the salivary glands, liver, pancreas, and gall bladder secrete/store enzymes/chemicals into the tract to facilitate digestion. The Digestive Process • In the mouth: english-lss.com – Food is partly broken down by the process of chewing and the chemical action of the enzymes secreted by the salivary glands. – Once the chewing action is completed, the food bolus is swallowed into the esophagus. -

Digestive System and Nutrition - Chapter 26
Digestive System and Nutrition - Chapter 26 All animals are heterotrophs and need organic molecules as food. We need food to get nutrients and energy. Body Mass Index (BMI) is a measure to understand desirable body weight for a person of certain height. It is considered overweight if its value is above 25. Formula used to calculate BMI is: Body Mass Index = body weight in Kg / height in m2 Cnidarians and flat worms have incomplete digestive system with one opening mouth. Actually this cavity acts as a gastrovascular cavity and serves as both digestive and vascular system. Complete digestive tract Round worms evolved complete digestive system with both mouth and anus. It has a tube within tube design. Most animals including humans have a digestive tract and associated glands. Human Digestive System Humans have digestive tract with: mouth mouth cavity pharynx esophagus stomach duodenum small intestine large intestine anus. The associated glands are salivary glands, liver, and pancreas. Liver and Pancreas Liver: is the largest gland in body. It stores its waste bile in Gall Bladder. Bile is released into duodenum and helps in digestion of fats. Liver secretion, bile, does not have any enzymes in it. Liver absorbs excess glucose from blood and release it back when needed. Pancreas: is the 2nd largest gland in human body. It secretes pancreatic juice into duodenum. The juice changes the acidic food from stomach into slightly basic food. It is very rich in digestive enzymes and help in digestion of starch, fats, proteins and nucleic acids. 4-stages of food-processing Ingestion is the eating of food by an animal. -
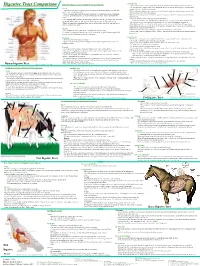
Digestive Tract Comparison • the Small Intestine Is a Tube Roughly Twenty Feet Long Deided Into the Duodenum, Jejunum and Ileum
• Small Intestine Human/Dog Digestive system or Simple Monogastric Digestion Digestive Tract Comparison • The small intestine is a tube roughly twenty feet long deided into the duodenum, jejunum and ileum. • The first part of the small intestine is the duodenum, the site of most chemical digestive reactions and is Mouth smoother than the rest of the small intestine • A specialized region of the digestive tract designed to break up large particles of food into • Bile, bicarbonate and pancreatic enzymes are secreted into the duodenum to breakdown nutrients in the smaller, more manageable particles chyme so that they can be readily absorbed. • Saliva is added to moisten food and begin carbohydrate breakdown by amylase in humans. •Bicarbonate from the pancreas neutralizes corrosive stomach acid from 3.5 in the stomach to 8.5 in the • There are four main types of teeth in the human or dog: incisors, canines, premolars and small intestine. molars. •Pancreatic enzymes include lipases, peptidases and amylases. •One reason dog and cat canines are much larger than ours is that they need to be able to rip and •Lipases break down fats. Peptidases break down proteins. Amylases break down carbohydrates. tear through tough raw meat. Humans have evolved to eat easier to chew, cooked meat. • Bile from the liver is stored in the gall bladder and secreted into the duodenum to emulsify fat. • While chewing, food is transformed into what is called a bolus, a food ball, and then forced •The jejunum and ileum are next in the small intestine and are covered in villi, finger-like projections. -
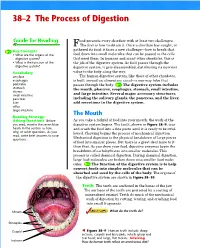
38–2 the Process of Digestion 1 FOCUS Objectives Ood Presents Every Chordate with at Least Two Challenges
Section 38–2 38–2 The Process of Digestion 1 FOCUS Objectives ood presents every chordate with at least two challenges. FThe first is how to obtain it. Once a chordate has caught, or 38.2.1 Identify the organs of the gathered its food, it faces a new challenge—how to break that digestive system. Key Concepts • What are the organs of the food down into small molecules that can be passed to the cells 38.2.2 Describe the function of the digestive system? that need them. In humans and many other chordates, this is digestive system. • What is the function of the the job of the digestive system. As food passes through the digestive system? digestive system, it gets disassembled, distributing its nutrient Vocabulary value to the body along the way. amylase The human digestive system, like those of other chordates, Vocabulary Preview esophagus is built around an alimentary canal—a one-way tube that peristalsis passes through the body. The digestive system includes If students have difficulty pronounc- stomach the mouth, pharynx, esophagus, stomach, small intestine, chyme ing any of the Vocabulary words, it and large intestine. Several major accessory structures, may interfere with their comprehen- small intestine pancreas including the salivary glands, the pancreas, and the liver, sion. Read each of the words to the liver add secretions to the digestive system. class and have students repeat them villus large intestine after you. Point out that villus is sin- The Mouth gular and the plural is villi. Reading Strategy: Asking Questions Before As you take a forkful of food into your mouth, the work of the Reading Strategy you read, rewrite the seven blue digestive system begins.