TRIM21 Polymorphisms Are Associated with Susceptibility And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cytosolic Fc Receptor TRIM21 Inhibits Seeded Tau Aggregation
Cytosolic Fc receptor TRIM21 inhibits seeded tau aggregation William A. McEwana,1, Benjamin Falcona, Marina Vaysburda, Dean Clifta, Adrian L. Oblakb, Bernardino Ghettib, Michel Goederta,1, and Leo C. Jamesa,1 aMedical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom; and bDepartment of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, IN 46202 Edited by Alexander Espinosa, Karolinska Institutet, Stockholm, Sweden, and accepted by Editorial Board Member Stephen P. Goff December 6, 2016 (received for review May 6, 2016) Alzheimer’s disease (AD) and other neurodegenerative disorders assemblies into the brains of tau transgenic mice promotes the are associated with the cytoplasmic aggregation of microtubule- aggregation of intracellular tau (7), and ectopic addition of associated protein tau. Recent evidence supports transcellular recombinant tau assemblies promotes the aggregation of in- transfer of tau misfolding (seeding) as the mechanism of spread tracellular pools in cultured cells (8, 9). Consistent with trans- within an affected brain, a process reminiscent of viral infection. cellular propagation of misfolding, tau pathology in AD spreads However, whereas microbial pathogens can be recognized as non- between connected regions of the brain in a stereotyped manner self by immune receptors, misfolded protein assemblies evade detec- (10). Seeded propagation of misfolded tau is therefore proposed tion, as they are host-derived. Here, we show that when misfolded to underlie many common neurodegenerative disorders. Akin to tau assemblies enter the cell, they can be detected and neutralized via viral infection, this model of seeded tau propagation relies on the a danger response mediated by tau-associated antibodies and the physical transfer of tau assemblies between cells, and is therefore cytosolic Fc receptor tripartite motif protein 21 (TRIM21). -

Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 As
cells Article Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 as New Binding Partners 1, 1, 2, Eunate Gallardo-Vara y, Lidia Ruiz-Llorente y, Juan Casado-Vela y , 3 4 5 6, , María J. Ruiz-Rodríguez , Natalia López-Andrés , Asit K. Pattnaik , Miguel Quintanilla z * 1, , and Carmelo Bernabeu z * 1 Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas (CSIC), and Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), 28040 Madrid, Spain; [email protected] (E.G.-V.); [email protected] (L.R.-L.) 2 Bioengineering and Aerospace Engineering Department, Universidad Carlos III and Centro de Investigación Biomédica en Red Enfermedades Neurodegenerativas (CIBERNED), Leganés, 28911 Madrid, Spain; [email protected] 3 Centro Nacional de Investigaciones Cardiovasculares (CNIC), 28029 Madrid, Spain; [email protected] 4 Cardiovascular Translational Research, Navarrabiomed, Complejo Hospitalario de Navarra (CHN), Universidad Pública de Navarra (UPNA), IdiSNA, 31008 Pamplona, Spain; [email protected] 5 School of Veterinary Medicine and Biomedical Sciences, and Nebraska Center for Virology, University of Nebraska-Lincoln, Lincoln, NE 68583, USA; [email protected] 6 Instituto de Investigaciones Biomédicas “Alberto Sols”, Consejo Superior de Investigaciones Científicas (CSIC), and Departamento de Bioquímica, Universidad Autónoma de Madrid (UAM), 28029 Madrid, Spain * Correspondence: [email protected] (M.Q.); [email protected] (C.B.) These authors contributed equally to this work. y Equal senior contribution. z Received: 7 August 2019; Accepted: 7 September 2019; Published: 13 September 2019 Abstract: Endoglin is a 180-kDa glycoprotein receptor primarily expressed by the vascular endothelium and involved in cardiovascular disease and cancer. -

A Study on the E3 Ligase TRIM21/Ro52 Alexander Espinosa from the Department of Medicine, Karolinska Institutet, Stockholm, Sweden
Thesis for doctoral degree (Ph.D.) 2008 Thesis for doctoral degree (Ph.D.) 2008 A STUDY ON THE E3 LIGASE TRIM21/RO52 A study on the E3 ligase TRIM21/Ro52 ligase E3 the on study A Alexander Espinosa Alexander Espinosa Fredrik Bredin From the Department of Medicine, Karolinska Institutet, Stockholm, Sweden A STUDY ON THE E3 LIGASE TRIM21/Ro52 Alexander Espinosa All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by Larserics Digital Print AB, Box 20082, SE-161 02 Bromma, Sweden. © Alexander Espinosa, 2008 ISBN 978-91-7357-480-8 iii ABSTRACT Patients with the systemic autoimmune diseases Sjögren's syndrome (SS) and systemic lupus erythematosus (SLE), often have antibodies against the intracellular protein TRIM21/Ro52. Although the presence of anti-TRIM21/Ro52 autoantibodies is used as a diagnostic tool, the biological function of TRIM21/Ro52 is still unknown. The aim of this thesis is to provide a better understanding of the function of TRIM21/Ro52, especially regarding its role in autoimmunity. To achieve this, TRIM21/Ro52 was studied at a molecular and cellular level in vitro and in vivo. By using circular dichroism, limited proteolysis, mass spectrometry, ultra-centrifugation, and two- hybrid experiments, it was shown that TRIM21/Ro52 is a Zn2+ binding protein that forms weak dimers. These results also confirmed the presence of the predicted secondary structure domains of TRIM21/Ro52. A RING domain in the N-terminus of TRIM21/Ro52 suggests that TRIM21/Ro52 is a RING dependent E3 ligase. Through ubiquitination assays, it was shown that TRIM/Ro52 is indeed an E3 ligase and that the E3 ligase activity of TRIM21/Ro52 was dependent on the E2 enzymes UbcH5a-c or UbcH6. -

Review Article Autoantigen TRIM21/Ro52 As a Possible Target for Treatment of Systemic Lupus Erythematosus
Hindawi Publishing Corporation International Journal of Rheumatology Volume 2012, Article ID 718237, 11 pages doi:10.1155/2012/718237 Review Article Autoantigen TRIM21/Ro52 as a Possible Target for Treatment of Systemic Lupus Erythematosus Ryusuke Yoshimi,1 Yoshiaki Ishigatsubo,1 and Keiko Ozato2 1 Department of Internal Medicine and Clinical Immunology, Yokohama City University Graduate School of Medicine, Yokohama 236-0004, Japan 2 Program in Genomics of Differentiation, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA Correspondence should be addressed to Ryusuke Yoshimi, [email protected] Received 11 January 2012; Revised 1 April 2012; Accepted 2 April 2012 Academic Editor: Javier Martin Copyright © 2012 Ryusuke Yoshimi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Systemic lupus erythematosus (SLE) is a chronic, systemic, and autoimmune disease, whose etiology is still unknown. Although there has been progress in the treatment of SLE through the use of glucocorticoid and immunosuppressive drugs, these drugs have limited efficacy and pose significant risks of toxicity. Moreover, prognosis of patients with SLE has remained difficult to assess. TRIM21/Ro52/SS-A1, a 52-kDa protein, is an autoantigen recognized by antibodies in sera of patients with SLE and Sjogren’s¨ syndrome (SS), another systemic autoimmune disease, and anti-TRIM21 antibodies have been used as a diagnostic marker for decades. TRIM21 belongs to the tripartite motif-containing (TRIM) super family, which has been found to play important roles in innate and acquired immunity. -
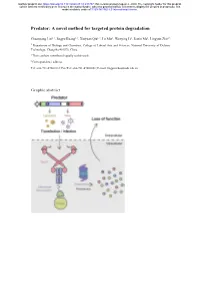
A Novel Method for Targeted Protein Degradation
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Predator: A novel method for targeted protein degradation Chuanyang Liu1, †, Jingyu Kuang1, †, Xinyuan Qiu1, †, Lu Min1, Wenying Li1, Jiaxin Ma1, Lingyun Zhu1,* 1 Department of Biology and Chemistry, College of Liberal Arts and Sciences, National University of Defense Technology, Changsha 410073, China † These authors contributed equally to this work. ∗ Correspondence address. Tel: +86-731-87001812; Fax/Tel: +86-731-87001801; E-mail: [email protected] Graphic abstract bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Protein expression and degradation are fundamental to cell function and physiological status of organisms. Interfering with protein expression not only provides powerful strategies to analyze the function of proteins but also inspires effective treatment methods for diseases caused by protein dysfunction. Recently, harnessing the power of the ubiquitin-proteasome system for targeted protein degradation (TPD) has become the focus of researches. Over the past two decades, TPD technologies, such as E3 ligase modification, PROTACs, and the Trim-Away method, have successfully re-oriented the ubiquitin-proteasome pathway and thus degraded many pathogenic proteins and even "undruggable" targets. -

S41467-019-12388-Y.Pdf
ARTICLE https://doi.org/10.1038/s41467-019-12388-y OPEN A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases Leo Kiss1,4, Jingwei Zeng 1,4, Claire F. Dickson1,2, Donna L. Mallery1, Ji-Chun Yang 1, Stephen H. McLaughlin 1, Andreas Boland1,3, David Neuhaus1,5 & Leo C. James1,5* 1234567890():,; The cytosolic antibody receptor TRIM21 possesses unique ubiquitination activity that drives broad-spectrum anti-pathogen targeting and underpins the protein depletion technology Trim-Away. This activity is dependent on formation of self-anchored, K63-linked ubiquitin chains by the heterodimeric E2 enzyme Ube2N/Ube2V2. Here we reveal how TRIM21 facilitates ubiquitin transfer and differentiates this E2 from other closely related enzymes. A tri-ionic motif provides optimally distributed anchor points that allow TRIM21 to wrap an Ube2N~Ub complex around its RING domain, locking the closed conformation and promoting ubiquitin discharge. Mutation of these anchor points inhibits ubiquitination with Ube2N/ Ube2V2, viral neutralization and immune signalling. We show that the same mechanism is employed by the anti-HIV restriction factor TRIM5 and identify spatially conserved ionic anchor points in other Ube2N-recruiting RING E3s. The tri-ionic motif is exclusively required for Ube2N but not Ube2D1 activity and provides a generic E2-specific catalysis mechanism for RING E3s. 1 Medical Research Council Laboratory of Molecular Biology, Cambridge, UK. 2Present address: University of New South Wales, Sydney, NSW, Australia. 3Present address: Department of Molecular Biology, Science III, University of Geneva, Geneva, Switzerland. 4These authors contributed equally: Leo Kiss, Jingwei Zeng. 5These authors jointly supervised: David Neuhaus, Leo C. -

TRIM21 Mediates Antibody Inhibition of Adenovirus-Based Gene Delivery
TRIM21 mediates antibody inhibition of adenovirus- based gene delivery and vaccination Maria Bottermanna, Stian Fossb,c,d, Laurens M. van Tienena, Marina Vaysburda, James Cruickshanka, Kevin O’Connella, Jessica Clarka, Keith Mayesa, Katie Higginsona, Jack C. Hirste, Martin B. McAdamc, Greg Slodkowicza, Edward Hutchinsone, Patrycja Kozika, Jan Terje Andersenb,c,d, and Leo C. Jamesa,1 aProtein and Nucleic Acid Chemistry Division, Medical Research Council, Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom; bDepartment of Biosciences, Centre for Immune Regulation, University of Oslo, N-0316 Oslo, Norway; cDepartment of Immunology, Centre for Immune Regulation, Oslo University Hospital, N-0372 Oslo, Norway; dDepartment of Pharmacology, Institute of Clinical Medicine, University of Oslo, N-0372 Oslo, Norway; and eMRC-University of Glasgow Centre for Virus Research, Glasgow G61 1QH, United Kingdom Edited by Michael B. A. Oldstone, The Scripps Research Institute, La Jolla, CA, and approved August 14, 2018 (received for review April 13, 2018) Adenovirus has enormous potential as a gene-therapy vector, but after endocytosis and can therefore carry antibody into the cy- preexisting immunity limits its widespread application. What is tosol. Upon binding to the Fc region of IgG with its PRYSPRY responsible for this immune block is unclear because antibodies motif, TRIM21 becomes activated and sequentially recruits the potently inhibit transgene expression without impeding gene E2 enzymes Ube2W and Ube2N/Ube2V2, which N-terminally transfer into target cells. Here we show that antibody prevention polyubiquitinate TRIM21 with anchored K63 chains. This results of adenoviral gene delivery in vivo is mediated by the cytosolic in recruitment of the proteasome and rapid degradation of the antibody receptor TRIM21. -

TRIM21 Monoclonal ANTIBODY Catalog Number:67136-1-Ig
For Research Use Only TRIM21 Monoclonal ANTIBODY www.ptglab.com Catalog Number:67136-1-Ig Catalog Number: GenBank Accession Number: Purification Method: Basic Information 67136-1-Ig BC010861 Protein A purification Size: GeneID (NCBI): CloneNo.: 150UL , Concentration: 1000 μg/ml by 6737 1E8B9 Bradford method using BSA as the Full Name: Recommended Dilutions: standard; tripartite motif-containing 21 WB 1:2000-1:10000 Source: Calculated MW: IHC 1:250-1:1000 Mouse 475 aa, 54 kDa Isotype: Observed MW: IgG1 52 kDa Immunogen Catalog Number: AG28377 Applications Tested Applications: Positive Controls: IHC, WB, ELISA WB : HeLa cells, Jurkat cells, HL-60 cells, HSC-T6 cells, Species Specificity: NIH/3T3 cells Human, mouse, rat IHC : human breast cancer tissue, human pancreas Note-IHC: suggested antigen retrieval with cancer tissue TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 TRIM21 has multiple N-terminal zinc finger motifs(has E3 ligase activity), a central leucine zipper, and a potential Background Information N-glycosylation site. TRIM3 is a E3 ubiquitin-protein ligase. It can form a ubiquitin ligase complex with E2 UBE2D2, which function not only for the uniquitination of USP4 and IKBKB but also for its self-ubiquitination. It has a role in the regulation of the cell cycle progression and could enhance the decapping activity of DCP2. It can exist in all mammalian cells as a ribonucleoprotein particle , which composed of a single polypeptide and 1 of 4 small RNA molecules. TRIM21 exists various isoforms and range molecular weight of isoforms is about 42-45kDa and 49-54 kDa. -

Rare Missense Variants in the Human Cytosolic Antibody Receptor Preserve Antiviral Function Jingwei Zeng†, Greg Slodkowicz†, Leo C James*
RESEARCH ARTICLE Rare missense variants in the human cytosolic antibody receptor preserve antiviral function Jingwei Zeng†, Greg Slodkowicz†, Leo C James* MRC Laboratory of Molecular Biology, Cambridge, United Kingdom Abstract The genetic basis of most human disease cannot be explained by common variants. One solution to this ‘missing heritability problem’ may be rare missense variants, which are individually scarce but collectively abundant. However, the phenotypic impact of rare variants is under-appreciated as gene function is normally studied in the context of a single ‘wild-type’ sequence. Here, we explore the impact of naturally occurring missense variants in the human population on the cytosolic antibody receptor TRIM21, using volunteer cells with variant haplotypes, CRISPR gene editing and functional reconstitution. In combination with data from a panel of computational predictors, the results suggest that protein robustness and purifying selection ensure that function is remarkably well-maintained despite coding variation. DOI: https://doi.org/10.7554/eLife.48339.001 Introduction Rare missense variants outnumber common ones, with 85% of non-synonymous variants displaying a minor allele frequency of less than 0.5% (Abecasis et al., 2012), and 200–300 such alleles per *For correspondence: sequenced individual (Bamshad et al., 2011). As each specific variant is present at a very low fre- [email protected] quency within the population, the impact on human health is hard to assess. Collectively however, † These authors contributed rare variants are thought to be a significant component of the ‘missing heritability’ paradigm and equally to this work their neglected contribution may explain why only a fraction of inherited diseases are genetically Competing interests: The accounted for Maher (2008). -

Inhibition of USP4 Attenuates Pathological Scarring by Downregulation of the TGF‑Β/Smad Signaling Pathway
MOLECULAR MEDICINE REPORTS 20: 1429-1435, 2019 Inhibition of USP4 attenuates pathological scarring by downregulation of the TGF‑β/Smad signaling pathway JIE ZHANG, SIJIA NA, SHUTING PAN, SANG LONG, YUQI XIN, QINGKUN JIANG, ZHONGWEI LAI, JUNFENG YAN and ZHONGYI CAO Department of Oral and Maxillofacial Surgery, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi 330006, P.R. China Received October 16, 2018; Accepted April 17, 2019 DOI: 10.3892/mmr.2019.10370 Abstract. Pathological scarring is a result of the hypertrophy Introduction of scar tissue during tissue repair following trauma. The aim of the present study was to assess the effect of ubiquitin‑specific Scars are the products of wound healing by the human protease 4 (USP4) silencing on pathological scarring, and to body. Among them, hypertrophic scars (HS) and keloids evaluate the mechanistic basis for the effect. An MTT assay are known as pathological scars, which are characterized by was used to assess cell viability. Immunoprecipitation (IP) was sustained hyperactive growth, irregular scar shape, and an used to determine ubiquitination levels of the TGF-β receptor uneven and tough texture (1). Pathological scars are the result (TβR)I and Smad7. Tumor formation was assessed by injecting of hypertrophy of scar tissue in the process of tissue repair keloid fibroblasts. Hematoxylin and eosin staining was used to following trauma. Pathological scars have serious aesthetic detect pathological changes in tumor tissue. Reverse transcrip- effects and result in tissue dysfunction due to deformities tion quantitative polymerase chain reaction and western blot of the scar contractures, particularly at the joints. Certain analysis assays were used to evaluate the expression levels of patients experience pain, itching, ulceration and the develop- TβRI and Smad7. -

Autoantigen TRIM21/Ro52 Is Expressed on the Surface of Antigen-Presenting Cells and Its Enhanced Expression in Sjögren's Synd
Sjögren syndrome RMD Open: first published as 10.1136/rmdopen-2020-001184 on 15 June 2020. Downloaded from SHORT REPORT Autoantigen TRIM21/Ro52 is expressed on the surface of antigen-presenting cells and its enhanced expression in Sjögren’s syndrome is associated with B cell hyperactivity and type I interferon activity Maarten R Hillen ,1,2 Katia Urso,3 Emma Koppe,3 Ana Pinheiro Lopes,1,2 Sofie L M Blokland,1,2 Aridaman Pandit ,1,2 Tom Slocombe,3 André van Maurik,4 Joel A G van Roon,1,2 Timothy R D J Radstake1,3 To cite: Hillen MR, Urso K, Tripartite motif-containing protein 21 et al Key messages Koppe E, . Autoantigen (TRIM21)/Ro52 is an immunoglobulin TRIM21/Ro52 is expressed on the surface of antigen- receptor that together with Ro60 makes up What is already known on this subject ’ presenting cells and its Sjögren s syndrome-antigen A (SSA). ► TRIM21/Ro52 is a major autoantibody target in enhanced expression in A 60%–80% of patients with primary Sjög- pSS and regulates downstream signalling of type ’ Sjögren ssyndromeis ren’s syndrome (pSS) have anti-SSA autoanti- IIFNs. associated with B cell bodies and they form part of the pSS ► Apoptosis-associated surface expression of TRIM21 hyperactivity and type TRIM21 on non-immune cells is associated with enhanced Iinterferonactivity.RMD Open diagnosis. is a cytosolic Fc-receptor 2020;6:e001184. doi:10.1136/ that mediates neutralisation of opsonised binding of anti-Ro52 autoantibodies. rmdopen-2020-001184 viruses and other particles, acting as a last What this study adds http://rmdopen.bmj.com/ line of defence against viruses that are recog- ► TRIM21 ► Additional material is Enhanced expression of in type I IFN- published online only. -

Overexpression of TRIB2 in Human Lung Cancers Contributes to Tumorigenesis Through Downregulation of C/Ebpa
Oncogene (2011) 30, 3328–3335 & 2011 Macmillan Publishers Limited All rights reserved 0950-9232/11 www.nature.com/onc ORIGINAL ARTICLE Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPa KB Grandinetti1,2,5, TA Stevens1,5,SHa1, RJ Salamone1, JR Walker1, J Zhang1, S Agarwalla1, DG Tenen3,4, EC Peters1 and VA Reddy1,6 1Genomics Institute of the Novartis Research Foundation, San Diego, CA, USA; 2Scripps Research Institute, San Diego, CA, USA; 3Cancer Science Institute, National University of Singapore and 4Harvard Stem Cell Institute, Center for Life Sciences, Boston, MA, USA Lung cancer is the most common cause of cancer-related et al., 2008; Schatton et al., 2008). As such, much mortality worldwide. Here, we report elevated expression research has gone into uncovering the different popula- of tribbles homolog 2 (TRIB2) in primary human lung tions of progenitor cells within the lung. These are tumors and in non-small cell lung cancer cells that express considered to be a rare population of undifferentiated low levels of differentiation-inducing transcription factor cells with enhanced ability to proliferate, self-renew, CCAAT/enhancer-binding protein alpha (C/EBPa). In and eventually differentiate into phenotypically diverse approximately 10–20% of cases, elevated TRIB2 expres- and heterogeneous cell populations (Kim et al., 2005). sion resulted from gene amplification. TRIB2 knockdown The CCAAT/enhancer-binding protein (C/EBP) family was found to inhibit cell proliferation and in vivo tumor of transcription factors is known to affect different aspects growth. In addition, TRIB2 knockdown led to morpholo- of development and differentiation in the lung (Cassel and gical changes similar to C/EBPa overexpression and Nord 2003).