Prevalence of Pathotypes of Phakopsora Pachyrhizi Syd. Causing Asian Soybean Rust in India
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

HŒ臬 A„簧綟糜恥sµ, Vw笑n® 22.12.2019 Š U拳 W
||Om Shri Manjunathaya Namah || Shri Kshethra Dhamasthala Rural Development Project B.C. Trust ® Head Office Dharmasthala HŒ¯å A„®ãtÁS®¢Sµ, vw¯ºN® 22.12.2019 Š®0u®± w®lµu® îµ±°ªæX¯Š®N®/ N®Zµ°‹ š®œ¯‡®±N®/w®S®u®± š®œ¯‡®±N® œ®±uµÛ‡®± wµ°Š® wµ°î®±N¯r‡®± ªRq® y®‹°£µ‡®± y®ªq¯ºý® D Nµ¡®w®ºruµ. Cu®Š®ªå 50 î®±q®±Ù 50 Oʺq® œµX®±Ï AºN® y®lµu®î®Š®w®±Ý (¬šµ¶g¬w®ªå r¢›Š®±î®ºqµ N®Zµ°‹/w®S®u®± š®œ¯‡®±N® œ®±uµÛSµ N®xÇ®Õ ïu¯ãœ®Áqµ y®u®ï î®±q®±Ù ®±š®±é 01.12.2019 NµÊ Aw®æ‡®±î¯S®±î®ºqµ 25 î®Ç®Á ï±°Š®u®ºqµ î®±q®±Ù îµ±ªæX¯Š®N® œ®±uµÛSµ N®xÇ®Õ Hš¬.Hš¬.HŒ¬.› /z.‡®±±.› ïu¯ãœ®Áqµ‡µ²ºvSµ 3 î®Ç®Áu® Nµ©š®u® Aw®±„Â®î® î®±q®±Ù ®±š®±é 01.12.2019 NµÊ Aw®æ‡®±î¯S®±î®ºqµ 30 î®Ç®Á ï±°Š®u®ºqµ ) î®±±ºvw® œ®ºq®u® š®ºu®ý®Áw®NµÊ B‡µ±Ê ¯l®Œ¯S®±î®¼u®±. š®ºu®ý®Áw®u® š®Ú¡® î®±q®±Ù vw¯ºN®î®w®±Ý y®äqµã°N®î¯T Hš¬.Hº.Hš¬ î®±²©N® ¯Ÿr x°l®Œ¯S®±î®¼u®±. œ¯cŠ¯u® HŒ¯å A„®ãtÁS®¢Sµ A†Ãw®ºu®wµS®¡®±. Written test Sl No Name Address Taluk District mark Exam Centre out off 100 11 th ward near police station 1 A Ashwini Hospete Bellary 33 Bellary kampli 2 Abbana Durugappa Nanyapura HB hally Bellary 53 Bellary 'Sri Devi Krupa ' B.S.N.L 2nd 3 Abha Shrutee stage, Near RTO, Satyamangala, Hassan Hassan 42 Hassan Hassan. -

District Census Handbook, Dharwad, Part XII-A, Series-11
CENSUS OF INDIA 1991 Series -11 KARNATAKA DISTRICT CENSUS HANDBOOK DHAR\VAD DISTRICT PART XII-A "IU.AGE ANHTOWN DIRELJORY SOBIIA NAMBISAN DH-ector of Census Operations, Kurnataka CONTENTS Page No. FOREWORD vii-viii PREFACE lX-X IMPORTANT STATISTICS ANALYTICAL NOTE Section-I - VHl&lge DiI'cctm'Y Explanatory Notc Alphabelical List of Villages - Bpdgi C.O.Blm:k Village Directory Statemcnt - B).tdgi C.D.Blud. Alphabetical List of Villages - Dhafwad CoD. Rlock Village Dircctory Statemcnt - Dharwad C.D.Block Alphahctical Liht of Villagch - (jadag C.O.BlI)(:k Village Directory Statement - Gadag CD.Block ()X- 105 Alphabetical Lihl of Villages - Hallgal C.D.Bhld: )1)1)- I 12 Village Directory Statement - Hangal CD.Block 11-1-1-11 Alphabetical Liht of Villages - Ha .... eri C.D.Block 145-147 Village Directory Stah:mcnl - 11a\L~ri C.D.Blod. 1-1X- J(,.1 Alphabetical Lihl of Vilbgl.: .... - } lirdcr lit' C.D.Block 1(,7-170 Village Directory Slal<.:m..:nl - I-lird. ..:rur C.D.BhK'1-.. 11'2-1:-;1) Alphabetical List of Villageh - lillbli C.D.BhlCk 1'J.1-194 ViI1age Directory Slat<.:ment - Hubli C.D.Block 1')(>-205 Alphabetical Liht of Villages - Kalg:h;tlgi C.D.Block 2()')- 21 I Village Dircctory Statcment - K4Ilghatgi CO.Block 212-225 Alphabclit'al List of Villages - K lInd;I~()1 CD.Block 22()-23() Village Directory Stat<.:ml'nt - Kundagul C.D.Block Alphabetical List of Villageh - MlInd,lrgi CD.B1o,:h Village Directory Statcl1ll:nt - l\lulllbrgi CO.Blnd P"g_L' l'< ll. Alphahctical Li~t 01" Villages - N :1I·g.und C [). -

All India Jain Youth Federation's Mahaveer Limb Centre, Hubli
All India Jain Youth Federation's Mahaveer Limb Centre, Hubli List of Beneficiaries Sl No Name of Complete Age Male / Income Type of Date on Total cost Subsidy Travel Boarding Surgical Total Number Escort Aadhaar Card Mobile Beneficiary Address Female aid which of aid provided cost and correctio of Days No Number (given) given lodging n 1 Shambu A/P Dhandapur 7 Male 7000 KAFO 04/04/201 3500 0 0 0 0 0 1 Yes 483217886358 8545084474 Yallappa TA, Naragund. Caliper 8 Kadachi. Dist. Gadag. 2 Makbulamhad A/PMundagod. 8 Male 6500 KAFO 02/04/201 3000 0 0 0 0 0 1 Yes 996011514777 9686081039 Nazeerahama Dist.Karwar Caliper 8 d Malligar 3 Shilpa A/PBevur. 33 Female 7000 KAFO 02/04/201 3500 0 0 0 0 0 1 Yes 000000000000 9743105199 Sadashiv TA/Dist. Caliper 8 Hosalli. Bagalakot 4 Basappa. A/PKulageri. Ta. 53 Male 6000 Below 02/04/201 3500 0 0 0 0 0 1 Yes 000000000000 7090584217 Hullappa. Kustagi. Dist. Knee 8 Walikar. Koppal. 5 Madhavasa. 27Shanti Talkise 43 Male 7000 Above 02/04/201 3500 0 0 0 0 0 1 Yes 765514930654 9945425295 Laxmansa. masari Road Knee 8 Pawar. Gadag. 6 S. Azadnagar 59 Female 6000 Below 02/04/201 3500 0 0 0 0 0 1 Yes 962856450881 9480009751 Thangeshwari Hospet Ballary Knee 8 . 7 Amrappa. A/P Sasavihal Ta, 62 Male 6500 Below 02/04/201 3500 0 0 0 0 0 1 Yes 964883210260 9611973011 Hanamappa. Kustagi. Knee 8 Korli. Dist,Koppal 8 Sunita. Singh Gan Painapur 40 Female 8000 Below 02/04/201 3500 0 0 0 0 0 1 Yes 470930508788 7744833357 SujanSingh Aligad [ U P] Knee 8 9 Doddappa. -

Bcs Annexure B
Particulars of Banking outlets- BCs Annexure B Quarter ended ----- District Contact SR Name Block Locality/Village Bank Name Base Branch Name Name Of the BC Number of BC Remarks 1 DHARWAD DHARWAD MARADGI KVGB SHIVALLI HASANSAB K TAHASILDAR 9916584615 2 DHARWAD DHARWAD HONNAPURA KVGB ARAWATGI SACHIN B KHADABADI 9686929452 3 DHARWAD DHARWAD MADANABAVI KVGB BRANCH OPENED 4 DHARWAD DHARWAD KOTABAGI KVGB BRANCH OPENED 5 DHARWAD DHARWAD HANGARKI KVGB 6 DHARWAD DHARWAD CHIKKAMALLIGEWADA SYNDICATE DHARWAD MAIN B M METYAL 8951693518 7 DHARWAD DHARWAD KYARAKOPPA SYNDICATE DHARWAD MAIN SHIVANAND S/O NINGAPPA 8088229068 8 DHARWAD DHARWAD HAROBELAVADI SYNDICATE DHARWAD MAIN MS. GEETA D/O BASAPPA TORANGATTI 9902867498 9 DHARWAD DHARWAD KARADIGUDDA SYNDICATE DHARWAD MAIN MAHABALESHWARA GANGAPPA G 9632372020 10 DHARWAD DHARWAD MAREWADA SYNDICATE DHARWAD MAIN GANGADHAR GURUSHANTAPPA H 9902224652 11 DHARWAD DHARWAD KURUBAGATTI SYNDICATE NARENDRA MS. GIRIJAVVA VEERABASANGOUDA PATIL 9902673705 12 DHARWAD DHARWAD YADAWAD SYNDICATE NARENDRA SMT. SHASHIKALA BASAVARAJ GALAGI 9986254676 13 DHARWAD DHARWAD KABBENUR ING VYSYA UPPINBETAGERI NINGAPPA 9742368145 14 DHARWAD DHARWAD KALLURA ING VYSYA UPPINBETAGERI SMT PARAVVA GUDAGATTI 9886760837 15 DHARWAD DHARWAD PUDAKALAKATTI ING VYSYA UPPINBETAGERI SMT. NEELAVVA CHANDRAYYA 9980695956 16 DHARWAD DHARWAD LOKUR ING VYSYA UPPINBETAGERI KAREBASAYYA VIRAKTIMATH 9901705105 17 DHARWAD DHARWAD BELUR VIJAYA Kotur Ravi B Sandure 9731150446 18 DHARWAD DHARWAD MANASUR VIJAYA Mangundi Karevva S Yarihakal 9916311130 19 DHARWAD -

Details of Technical Experts and Farmers
Details of technical experts of UAS, Dharwad campus Sl. No. Name of the Scientist Expertise Contact No. 1. Dr. Angadi S. S. Cropping system 9449065459 2. Dr. Basavaraj Banakar Agriculture marketing 9448632398 3. Dr. M. V. Manjunatha Soil and water conservation 9449063372 4. Dr. Satish R. Desai Farm power and machineries 9448495341 5. Dr. C. B. Meti Soil and water conservation engineering 9449419613 6. Dr. Anuraj Farm power and machineries 9731907477 7. Dr. R. R. Patil Sericulture 9448894084 8. Dr. G. M. Patil Sericulture 9480461296 9. Dr. Bheemappa A. Agricultural extension 9449121372 10. Dr. S. S. Dolli Agricultural extension 9845142796 11. Dr. M. N. Sreenivasa Agriculture microbiology 9448220214 12. Dr. K. S. Jagadish Agriculture microbiology 9972755112 13. Dr. S. V. Hosmani Fodder production 9448497359 14. Dr. J. A. Mulla Poultry science 9448861650 15. Dr. S. D. Kololgi Dairy technology 9448497350 16. Dr. Ramesh Bhat Biotechnology 9945667300 17. Dr. Geeta G. Shirnalli Biogas production 9980024958 18. Dr. S. K. Deshpande Pulse breeder 9448214978 19. Dr. P. R. Dharamatti Vegetable breeder 9449070811 20. Dr. Venugopal C. K. Medicinal and aromatic plants 9448630255 21. Dr. R. V. Hegde Spices and plantation crops 9448923184 22. Dr. V. B. Nargund Mycology 9448797254 23. Dr. A. S. Byadgi Virology 9343104551 24. Dr. B. I. Bidari Soil Science & agril. chemistry 9449064306 25. Dr. Manjunath Hebbar Soil Science & agril. chemistry 9342218691 26. Dr. V. B. Kuligod Soil Science & agril. chemistry 9343167764 27. Dr. Anil Kumar Mugali Veterinary science 9845708013 28. Dr. Anil S. Patil Veterinary science 9731088592 29. Dr. Renuka S. Salunke Family resource management 9900484680 30. Dr. -

S Name Address 2010.Xls
Name and Address of the BLO's Name of Assembly Constituency: 70-Kundgol Total No. of Parts in the AC: 163 Total No. of BLO's in the AC: 163 District: Dharwad Desgination Complete Address of Part No. Name of the BLO's Contact No. ( T / NT) the BLO's 1 2 3 4 5 Annapurna Kumbar NT At: Revadihal Ta: Hubli 1 Anganavadi Worker 9611381737 Sarojini Ballari NT At: Devaragudihal Ta:Hubli 2 Anganavadi Worker 9731617821 Kamalaxi Sahadevappa Arji NT At: Parasapur Ta: Hubli 3 Anganavadi Worker 9036409754 Anasuyya S Hosur NT At: Rayanal Ta: Hubli 4 Anganavadi Worker 9739570040 Manjunath M Madlingannavar NT At: Rayanal Ta: Hubli 5 9448786060 Gram Panchayat Clerk Channamma B Sirigeri NT At: Gangival Ta:Hubli 6 Anganavadi Worker 9902663988 Girijamma I Budnal NT At:Anchatageri Ta:Hubli 7 Anganavadi Worker PP-9972991196 Bhagyalaxami A Kambalagi NT At:Anchatageri Ta:Hubli Anganavadi Worker 8 9986170419 K. M. Shashikala Teacher At:A Timmasagar Ta:Hubli 9 Asst Master 9986971331 Kasturi Kamaladinni NT At:A Mavanur Ta:Hubli 10 Anganavadi Worker 9844413915 Kasturibai Sudi NT At:A Mavanur Ta:Hubli 11 Anganavadi Worker 9632426177 Shailaja V Kulakarni At:A Kotagondhunshi NT 12 Anganavadi Worker Ta:Hubli (0836) 2317160 Mahadevi Totad NT At: Kadapatti Ta: Kundgol 13 Anganavadi Worker 9916122906 S S Jadimath NT At: Allapur Ta: Kundgol 14 Anganavadi Worker 9986814192 V K Badiger Teacher At: Gudenakatti Ta:Kundgol 15 Asst Master 9008628870 Yashoda Nagappa Arkasali( At:Benakanahalli Badiger NT 16 Ta:Kunadgol 9845698944 Anganavadi Worker Indira D Topanagoudar NT At: Mullolli Ta:Kundgol 17 Anganavadi Worker 9972325606 Dyamavva Chalawadi At: Yarinarayanpur NT 18 Anganavadi Worker Ta:Kundgol 9481730761 Sharada M Shalavadi NT At:Yaraguppi Ta:Kudngol 19 Anganavadi Worker 08304-265316 Annapurna Vijayakumar Madivalar NT At: Yaraguppi Ta:Kudngol 20 Anganavadi Worker 9844202516 Page 1 of 8 Desgination Complete Address of Part No. -

27.06.2013 1. Since Two Petitioners' Sought Information Is on the Same
KIC 2892 PTN 2013 Cw KIC 2897 PTN 2013 KARNATAKA INFORMATION COMMISSION, COURT HALL No.6 No.14/3, First Floor, Sri Aravind Bhavan [Mythic Society], Nrupatunga Road, Bangalore 560 001. Commission Website address http://www.kic.gov.in [Sri Gurusiddappa and Sri G. G. Ainapurmath vs. Secretary/PDO & PIO, Grama Panchayath Rayanal, Hubli Taluk, Dharwad District] O R D E R 27.06.2013 1. Since two Petitioners’ sought information is on the same subject and Respondents are same, hence both complaints are clubbed together and heard. 2. Petitioner is absent. Sri A. C. Chadragiri, Secretary & PIO, Grama Panchayath Rayanal, Hubli Taluk, Dharwad District is present along with Smt. Ansuya Sunkad, PDO. 3. Petitioners’ in his two requests for information dated 20.12.2012 & 26.10.2012 addressed to Secretary/PDO & PIO, Grama Panchayath Rayanal, Hubli Taluk, Dharwad District, has sought the following information: [2892] UÀAVªÁ¼À UÁæªÀÄzÀ j ¸ÀªÉð £ÀA 01/243J gÀ°è 63 ¥ÁèlÄUÀ¼À PÀÄjvÀÄ ºÁUÀÆ EvÀgÉ [2897] Regarding 4(1)(a) & 4(1)(b) 4. Petitioners have filed complaints to the Commission u/s 18(1) of the Act on 22.02.2013 & 25.02.2012, alleging that they have not received the information by the PIO for both applications. Subsequently, the Commission issued summons to all Parties to appear before the Commission on 27.06.2013 at 11.00 A.M. 5. Respondent Sri A. C. Chadragiri, Secretary & PIO states that, he is I/C of three Grama Panchayaths, hence because of the overload of the work he could not provided the information in time to the Petitioners and provided the information to the Petitioners on 27.06.2013, through RPAD and produced the copy of the Postal Acknowledgement for having sent the information. -
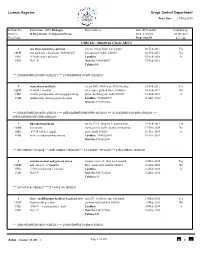
Dharwad Circle-ADC2
Licence Register Drugs Control Department Print Date : 31-Mar-2020 Sr/Firm No Firm Name , I.C / Manager Firm Address Issue Dt,Validity Cold Storage District / R.Pharmacist , Competent Person Dt,Renewal Dt, 24 Hr Open Firm Cons. Inspection Dt Lic App CIRCLE : Dharwad Circle-ADC2 1 m/s shree banashree pharma site no.190,gr.floor, narayanpur 03-Feb-2011 Yes 11151 smt parimala v kulakarni, 9880348607 oni,amargol, hubli. 580025 02-Feb-2021 No HB2 -52460-banu v kulkarni Landline. 03-Feb-2016 A PRO No C.P Mob No:9880348607 27-Feb-2016 Taluka:043 *** 20-KA/DWR/20/1397~02/02/21 *** 21-KA/DWR/21/1397~02/02/21 2 vigneshwar medicals cts.no 3007-3008, b.no.3105-06,shop 21-Feb-2011 Yes 11197 vinayak t. anaskar no.8, upper ground floor, madhura 20-Feb-2021 No HB2 -38043-pushpalatha shivayogappa shedg plaza, daziban peth, hubli 580028 21-Feb-2016 A PAR pushpalatha shivayogappa shedgali Landline. 9740606399 15-May-2017 Mob No:9972396062 *** 20-KA/DWR/20/1400~20/02/21 *** 20B-KA/DWR/20B/1059~20/02/21 *** 21-KA/DWR/21/1400~20/02/21 *** 21B-KA/DWR/21B/1008~20/02/21 3 bhavikatti medicals cts.no.172/1, shop no.5, ground floor, 19-Feb-2015 Yes 11203 b n swamy eureka tower traffic island chennamma 17-Dec-2024 No HB2 -49714-vivek i jugali circle hubli 580029 18-Dec-2019 A PAR sri b. neelakanteshwar swamy Landline. 9945525841 19-Dec-2019 Mob No:9945525841 *** 20-129280~17/12/24 *** 20B-129622~18/02/20 *** 21-129281~17/12/24 *** 21B-129623~18/02/20 4 renuka medical and general stores property no.1/a3, shop no.2 ground 16-Mar-2016 Yes 11224 smt. -

S.No Name of Grma Panchayat Identified
SLBC KARNATAKA: GRAM PANCHAYAT WISE SERVICE AREA PLAN OF THE DISTRICT: DHARWAR Name of the District: Dharwar Name Of the Block: BR./BC/A Name of Name of all revenue Post Identified TM Population Of the villages forming the GP Village Code Office/S Name of Grma place/village of Block existing Sl.N the Village as S.No allotted Used in 2011 ubpost Panchayat 2000+populatio Name or o. per Census bank with Census Office n(2001 census) proposed 2001 Branch YES/NO with name 1 AMMINBHAVI Dharwar BR 1 AMMINBHAVI 602299 10889 YES 2 ARAVATAGI Dharwar BC 2 AMBOLI 602248 1344 NO Dharwar BR 3 ARAVATAGI 602247 944 YES Dharwar BC 4 KOGILAGERI 602238 1204 NO Dharwar BC 5 KUMBARKOPPA 602250 1694 NO 3 BELUR BELUR Dharwar VIJAYA KOTUR BC 1 BEILUR 602226 2280 NO Dharwar BC 2 HEGGARI 602231 650 NO Dharwar BC 3 NEERALKATTI 602220 1251 NO 4 BENACHI Dharwar BC 1 BALAGERI 602238 1204 NO Dharwar BC 2 BENAGHI 602250 1696 YES Dharwar BC 3 DORI 602249 1696 NO 5 DEVARHUBLI Dharwar BC 1 DEVARHUBLI 602265 1954 NO Dharwar BC 2 MALLURA 602279 42 NO Dharwar BC 3 MURAKATTI 602264 822 NO 6 GARAG Dharwar BC 1 AGASANHALLI 602213 149 NO Dharwar BR 2 GARAG 602214 9422 YES 7 HALLIGERI Dharwar BC 1 AMLIKOPPA 602267 849 NO Dharwar BC 2 HALLIGERI 602266 1287 YES Dharwar BC 3 HULTIKOTI 602258 1134 NO Dharwar BC 4 MAVINKOPPA 602259 461 NO Dharwar BC 5 RAGIKALLAPUR NO 8 HANGARKI Dharwar BC 1 DUBBANAMARADI 620212 500 NO HANGARKI Dharwar KVGB TADAKOD BC 2 HANGARKI 602206 2003 NO Dharwar BC 3 KM TADAKOD 602199 695 NO Dharwar BC 4 SHEDABALA 602207 89 YES 9 HAROBELAVADI HAROBELAVADI Dharwar -
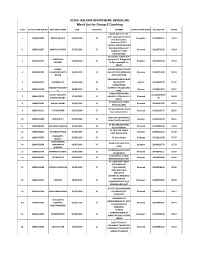
Merit List for Group-C Coaching
SOCIAL WELFARE DEPARTMENT, BENGALURU Merit List for Group-C Coaching SL NO APPLICATION REG NO APPLICANT NAME DOB CATEGORY ADDRESS EXAM CENTER NAME SSLC REG NO SCORE #6/19, 2nd cross, 4th main Jayanagar 1st block 1 1819G002896 PREETHAM M P 05/01/1994 SC Bengaluru 20100449134 92.04 east, Byrasandra, Bangalore-560011 C/O ER.S.H.RATHOD SHRI- RAM BUILDING VAJAY 2 1819G002837 AMRUTA RATHOD 27/07/1995 SC Dharwad 20110370342 90.24 NAGAR 1ST MAIN HALIYAL ROAD shreeshail y loganvi dist- SHREESHAIL vijayapur TQ- B.Bagewadi 3 1819G001395 01/06/1984 SC Belgaum 20030244284 89.33 LOGANVI At Post- manag0li Pin 586122 SURESH SURESH DASAR AT POST 4 1819G000338 YALAGURADDAPPA 25/09/1993 SC HULLUR TQ MUDDEBIHAL Dharwad 20100271498 88.29 DASAR DIST VIJAYAPUR BANASAMUDRAVILLAIGE 5 1819G003417 VINODRAJ B S 10/04/1996 SC TKHALLIPOST Mysore 20120539109 87.87 KASABAHOBLI SHARATH KUMAR H KOTHATHI VILLAGE AND 6 1819G003258 28/08/1995 SC Mysore 20110614504 87.11 P POST. RAMALINGESHWARA RAJEEV KOLLAPPA M11064375859 7 1819G001079 12/05/1995 SC NAGAR AT POST BELLATTI Dharwad 86.92 BANDIVADDARA 31 582112 AT VIRUPAPUR TANDA 8 1819G002501 ARJUN LAMANI 21/05/1993 SC Dharwad 20090387978 86.62 POST KALAKERI 5th ward behind masjid 9 1819G001631 Y TEJASHWINI 03/07/1996 SC Dharwad 20120109244 86.26 near railway station HOSA URU IBRAMPURA 10 1819G003485 NAGARAJA T 03/07/1996 SC Koppala 20120116647 86.11 VILAGE BAGEVADI POST AT DEVARAGUDIAHAL 11 1819G000032 MALLAPPA HARIJAN 21/02/1995 SC Dharwad 20110382399 85.89 POST RAYANAL AT CHATTAR TANDA 12 1819G003346 SITARAM RATHOD 07/06/1992 SC Koppala 20080161644 85.58 POST NAGALAPUR PRASHANT L 13 1819G003079 01/06/1995 SC AT post mashal Gulbarga 20120212468 85.50 JADHAV VIJAYAKUMAR dombar tot nipanal tq 14 1819G002968 YAMANAPPA 01/07/1992 SC Belgaum 20090319770 85.33 raybg DOMBAR AT NEERALAKATTI POST 15 1819G002787 SHANKAR AKKASALI 19/07/1988 SC Dharwad 20040340117 85.07 CHANDAPUR VIJAYAPURA VILLAGE 16 1819G000431 PRIYANKA K S 08/01/1991 SC Shimoga 20060610990 85.00 BENAKANAHALLI POST AT. -

District Census Handbook, Dharwad, Part XII-A & B, Series-30
CENSUS OF INDIA 2001 SERIES - 30 KARNATAKA PART XII - A & B : VILLAGE AND TOWN DIRECTORY & PRIMARY CENSUS ABSTRACT DISTRICT CENSUS HANDBOOK DHARWAD DIRECTORATE OF CENSUS OPERATIONS, KARNATAKA, BANGALORE Motif Unkal Tank near Hubli of Dharwad district Unkal Tank is located at a distance of 4 kms. from Unkal village is located at a distance of 4 kms., Hubli. Unkal Tank is one of the surviving tanks of from Hubli has a historical past. Unkal tank covers 250 Hubli City and will celeberate the centenary year dur acres with a catchment area of 4,651 Sq.Kms. A ing 2003. The then Municipal Authorities decided to number of migratory birds visits the tank. Due to rapid construct a tank near Unkal in 1886 in order to meet urbanization in the surrounding areas of the tank the the drinking water requirements of around 30,000 water supply has been stopped and declared unfit for people. Through the efforts of Great Statesman-Archi human consumption. Efforts to revive the past glory tect Sir.M.Vishweshwaraiah this project became reality for the tank is going on under National Tank Protection in 1903, it served as an important source of drinking Scheme water for the people of Hubli till the construction of Source : Deccan Herald Neerasagara and Malaprabha Reservoirs after Inde pendence. to 0 >. 0 ,., C\J ~ :0' @ • [EJ c .... ;:l ';::"" 0 >, ., ..Cl • I ::.:: 0 z ". u"" .I @ .3 I 00 ...." :a'" :5 ..s 'Ii '0 -' ~ <::: · • C Q) · E r.l 6 <::: · '- .... ;:0'-' Q) :> < I' '"0 0 '-' u '-' A N a< "" "' p:: >""a m co co Z < a "'" @ Z' ... -

Kalaghatagi Taluk Voters List S.No Reg No/MEM No Name & Address 1 Reg No.1167 Reg No.1167 Shri.G.M.Hiremath at & Post - B
All India Veerashaiva Mahasabha (R.) Bangalore Kalaghatagi Taluk Voters List S.No Reg No/MEM No Name & Address 1 Reg No.1167 Reg No.1167 Shri.G.M.Hiremath At & Post - B. Gudihala, Kalaghatagi Taluk, Dharwad Dist. 2 Reg No.1172 Reg No.1172 Shri.Ishwaragowda Linganagowda Patil Suresh Shetti, At & Post - Koppa,-581272 Kalaghatagi Taluk, Dharwad Dist. 3 Reg No.1174 Reg No.1174 Shri.C.M.Nimbennanavara At & Post - Mishrikoti, Kalaghatagi Taluk, Dharwad Dist. 4 Reg No.1173 Reg No.1173 Shri.Chanappa B.Honnihalli, Teacher, At & Post - Gandhinagar, Kalaghatagi Taluk, Dharwad Dist. 5 Reg No.2139 Reg No.2139 Shri.Chandrashekar Mahanthappa Thenginakai Akkipete, Kalaghatagi - 581204, Dharwad Dist. 6 Reg No.2153 Reg No.2153 Shri.Nigappa Ishwarappa Katti, Market Street, Kalaghatagi - 581204, Dharwad Dist. 7 Reg No.2154 Reg No.2154 Shri.Sangaiah V.Hiremath Gowdara Oni, Kalaghatagi - 581204, Dharwad Dist. 8 Reg No.2180 Reg No.2180 Shri.Mahadevappa Channappa Baligara Gowdara Oni, Kalaghatagi - 581204, Dharwad Dist. 9 Reg No.2181 Reg No.2181 Shri.Muthuraja T.Biradhar Patil Gowdara Oni, Kalaghatagi - 581204, Dharwad Dist. 10 Reg No.2182 Reg No.2182 Shri.Kallappa B.Meti At & Post - Madakihonnihalli, Kalaghatagi Taluk, Dharwad Dist. 11 Reg No.6932 Reg No.6932 Shri.Shakarapa Chanabasappa Angadi At & Post - Jinnura, Kalaghatagi Taluk, Dharwad Dist.- 58000. 12 Reg No.7124 Reg No.7124 Shri.Channappa Fakkirappa Swari At - Chikkanathar, Post - Hirenathar, Kalaghatagi Taluk, Dharwad Dist. 13 Reg No.7125 Reg No.7125 Shri.Shankarappa Ishwarappa Bolannavar At & Post - Begura - 581204, Kalaghatagi Taluk, Dharwad Dist. 14 Reg No.7126 Reg No.7126 Shri.Manjunath Chanappa Jyothi At & Post - Kudalagi - 581204, Kalaghatagi Taluk, Dharwad Dist.