Pydap Documentation Release 3.2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LAMP and the REST Architecture Step by Step Analysis of Best Practice
LAMP and the REST Architecture Step by step analysis of best practice Santiago Gala High Sierra Technology S.L.U. Minimalistic design using a Resource Oriented Architecture What is a Software Platform (Ray Ozzie ) ...a relevant and ubiquitous common service abstraction Creates value by leveraging participants (e- cosystem) Hardware developers (for OS level platforms) Software developers Content developers Purchasers Administrators Users Platform Evolution Early stage: not “good enough” solution differentiation, innovation, value flows Later: modular architecture, commoditiza- tion, cloning no premium, just speed to market and cost driven The platform effect - ossification, followed by cloning - is how Chris- tensen-style modularity comes to exist in the software industry. What begins as a value-laden proprietary platform becomes a replaceable component over time, and the most successful of these components finally define the units of exchange that power commodity networks. ( David Stutz ) Platform Evolution (II) Example: PostScript Adobe Apple LaserWriter Aldus Pagemaker Desktop Publishing Linotype imagesetters NeWS (Display PostScript) OS X standards (XSL-FO -> PDF, Scribus, OOo) Software Architecture ...an abstraction of the runtime elements of a software system during some phase of its oper- ation. A system may be composed of many lev- els of abstraction and many phases of opera- tion, each with its own software architecture. Roy Fielding (REST) What is Architecture? Way to partition a system in components -

A Learner's Guide to Programming Using the Python Language.Pdf
Advance Praise for Head First Programming “Head First Programming does a great job teaching programming using an iterative process. Add a little, explain a little, make the program a little better. This is how programming works in the real world and Head First Programming makes use of that in a teaching forum. I recommend this book to anyone who wants to start dabbling in programming but doesn’t know where to start. I’d also recommend this book to anyone not necessarily new to programming, but curious about Python. It’s a great intro to programming in general and programming Python specifically.” — Jeremy Jones, Coauthor of Python for Unix and Linux System Administration “David Griffiths and Paul Barry have crafted the latest gem in the Head First series. Do you use a computer, but are tired of always using someone else’s software? Is there something you wish your computer would do but wasn’t programmed for? In Head First Programming, you’ll learn how to write code and make your computer do things your way.” — Bill Mietelski, Software Engineer “Head First Programming provides a unique approach to a complex subject. The early chapters make excellent use of metaphors to introduce basic programming concepts used as a foundation for the rest of the book. This book has everything, from web development to graphical user interfaces and game programming.” — Doug Hellmann, Senior Software Engineer, Racemi “A good introduction to programming using one of the best languages around, Head First Programming uses a unique combination of visuals, puzzles, and exercises to teach programming in a way that is approachable and fun.” — Ted Leung, Principal Software Engineer, Sun Microsystems Praise for other Head First books “Kathy and Bert’s Head First Java transforms the printed page into the closest thing to a GUI you’ve ever seen. -

Openstack Security Guide April 26, 2014 Current
OpenStack Security Guide April 26, 2014 current OpenStack Security Guide current (2014-04-26) Copyright © 2013 OpenStack Foundation Some rights reserved. This book provides best practices and conceptual information about securing an OpenStack cloud. Except where otherwise noted, this document is licensed under Creative Commons Attribution 3.0 License. http://creativecommons.org/licenses/by/3.0/legalcode i TM OpenStack Security Guide April 26, 2014 current Table of Contents Preface ................................................................................................ 11 Conventions ................................................................................. 11 Document change history ............................................................ 11 1. Acknowledgments ............................................................................. 1 2. Why and how we wrote this book ..................................................... 3 Objectives ...................................................................................... 3 How .............................................................................................. 3 How to contribute to this book ..................................................... 7 3. Introduction to OpenStack ................................................................. 9 Cloud types ................................................................................... 9 OpenStack service overview ......................................................... 11 4. Security Boundaries and Threats -
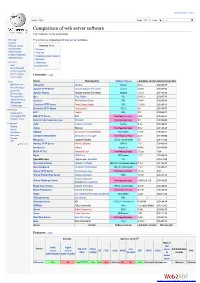
Comparison of Web Server Software from Wikipedia, the Free Encyclopedia
Create account Log in Article Talk Read Edit ViewM ohrisetory Search Comparison of web server software From Wikipedia, the free encyclopedia Main page This article is a comparison of web server software. Contents Featured content Contents [hide] Current events 1 Overview Random article 2 Features Donate to Wikipedia 3 Operating system support Wikimedia Shop 4 See also Interaction 5 References Help 6 External links About Wikipedia Community portal Recent changes Overview [edit] Contact page Tools Server Developed by Software license Last stable version Latest release date What links here AOLserver NaviSoft Mozilla 4.5.2 2012-09-19 Related changes Apache HTTP Server Apache Software Foundation Apache 2.4.10 2014-07-21 Upload file Special pages Apache Tomcat Apache Software Foundation Apache 7.0.53 2014-03-30 Permanent link Boa Paul Phillips GPL 0.94.13 2002-07-30 Page information Caudium The Caudium Group GPL 1.4.18 2012-02-24 Wikidata item Cite this page Cherokee HTTP Server Álvaro López Ortega GPL 1.2.103 2013-04-21 Hiawatha HTTP Server Hugo Leisink GPLv2 9.6 2014-06-01 Print/export Create a book HFS Rejetto GPL 2.2f 2009-02-17 Download as PDF IBM HTTP Server IBM Non-free proprietary 8.5.5 2013-06-14 Printable version Internet Information Services Microsoft Non-free proprietary 8.5 2013-09-09 Languages Jetty Eclipse Foundation Apache 9.1.4 2014-04-01 Čeština Jexus Bing Liu Non-free proprietary 5.5.2 2014-04-27 Galego Nederlands lighttpd Jan Kneschke (Incremental) BSD variant 1.4.35 2014-03-12 Português LiteSpeed Web Server LiteSpeed Technologies Non-free proprietary 4.2.3 2013-05-22 Русский Mongoose Cesanta Software GPLv2 / commercial 5.5 2014-10-28 中文 Edit links Monkey HTTP Server Monkey Software LGPLv2 1.5.1 2014-06-10 NaviServer Various Mozilla 1.1 4.99.6 2014-06-29 NCSA HTTPd Robert McCool Non-free proprietary 1.5.2a 1996 Nginx NGINX, Inc. -
![Arxiv:1903.01950V2 [Cs.OS] 20 Nov 2019](https://docslib.b-cdn.net/cover/1273/arxiv-1903-01950v2-cs-os-20-nov-2019-1791273.webp)
Arxiv:1903.01950V2 [Cs.OS] 20 Nov 2019
Pyronia: Intra-Process Access Control for IoT Applications Marcela S. Melara, David H. Liu, Michael J. Freedman Princeton University Abstract recognize_face() function could allow an attacker to steal the application’s private authentication token by upload Third-party code plays a critical role in IoT applications, this file to an unauthorized remote server. Meanwhile, the which generate and analyze highly privacy-sensitive data. application developer only expected recognize_face() to Unlike traditional desktop and server settings, IoT devices access the image file face.jpg. mostly run a dedicated, single application. As a result, This is an especially serious problem for IoT because vulnerabilities in third-party libraries within a process most devices run a single, dedicated application that has pose a much bigger threat than on traditional platforms. access to all sensors on the device. In other words, third- We present Pyronia, a fine-grained access control sys- party code does not run with least privilege [48]. tem for IoT applications written in high-level languages. Now, these threats are not IoT-specific. A large body of Pyronia exploits developers’ coarse-grained expectations prior research has sought to restrict untrusted third-party about how imported third-party code operates to restrict code in desktop, mobile, and cloud applications (e.g., [10, access to files, devices, and specific network destinations, 11,14,19,21,24,36,41,54,55,59,65 –67]). However, these at the granularity of individual functions. To efficiently traditional compute settings face more complex security protect such sensitive OS resources, Pyronia combines challenges. Mobile devices, desktops, and even IoT hubs, three techniques: system call interposition, stack inspec- run multiple mutually distrusting applications; further, tion, and memory domains. -

Chaussette Documentation Release 1.3.0
Chaussette Documentation Release 1.3.0 Tarek Ziade Sep 27, 2017 Contents 1 Usage 3 1.1 Running a plain WSGI application....................................3 1.2 Running a Django application......................................3 1.3 Running a Python Paste application...................................3 2 Using Chaussette in Circus 5 3 Using Chaussette in Supervisor7 4 Backends 9 5 Rationale and Design 11 6 Useful links 13 i ii Chaussette Documentation, Release 1.3.0 Chaussette is a WSGI server you can use to run your Python WSGI applications. The particularity of Chaussette is that it can either bind a socket on a port like any other server does or run against already opened sockets. That makes Chaussette the best companion to run a WSGI or Django stack under a process and socket manager, such as Circus or Supervisor. Contents 1 Chaussette Documentation, Release 1.3.0 2 Contents CHAPTER 1 Usage You can run a plain WSGI application, a Django application, or a Paste application. To get all options, just run chaussette –help. Running a plain WSGI application Chaussette provides a console script you can launch against a WSGI application, like any WSGI server out there: $ chaussette mypackage.myapp Application is <function myapp at 0x104d97668> Serving on localhost:8080 Using <class chaussette.backend._wsgiref.ChaussetteServer at 0x104e58d50> as a backend Running a Django application Chaussette allows you to run a Django project. You just need to provide the Python import path of the WSGI application, commonly located in the Django project’s wsgi.py file. For further information about how the wsgi. py file should look like see the Django documentation. -

Red Hat Enterprise Linux 7 7.8 Release Notes
Red Hat Enterprise Linux 7 7.8 Release Notes Release Notes for Red Hat Enterprise Linux 7.8 Last Updated: 2021-03-02 Red Hat Enterprise Linux 7 7.8 Release Notes Release Notes for Red Hat Enterprise Linux 7.8 Legal Notice Copyright © 2021 Red Hat, Inc. The text of and illustrations in this document are licensed by Red Hat under a Creative Commons Attribution–Share Alike 3.0 Unported license ("CC-BY-SA"). An explanation of CC-BY-SA is available at http://creativecommons.org/licenses/by-sa/3.0/ . In accordance with CC-BY-SA, if you distribute this document or an adaptation of it, you must provide the URL for the original version. Red Hat, as the licensor of this document, waives the right to enforce, and agrees not to assert, Section 4d of CC-BY-SA to the fullest extent permitted by applicable law. Red Hat, Red Hat Enterprise Linux, the Shadowman logo, the Red Hat logo, JBoss, OpenShift, Fedora, the Infinity logo, and RHCE are trademarks of Red Hat, Inc., registered in the United States and other countries. Linux ® is the registered trademark of Linus Torvalds in the United States and other countries. Java ® is a registered trademark of Oracle and/or its affiliates. XFS ® is a trademark of Silicon Graphics International Corp. or its subsidiaries in the United States and/or other countries. MySQL ® is a registered trademark of MySQL AB in the United States, the European Union and other countries. Node.js ® is an official trademark of Joyent. Red Hat is not formally related to or endorsed by the official Joyent Node.js open source or commercial project. -

Red Hat Enterprise Linux 7 Migration Planning Guide
Red Hat Enterprise Linux 7 Migration Planning Guide Migrating to Red Hat Enterprise Linux 7 Laura Bailey Red Hat Enterprise Linux 7 Migration Planning Guide Migrating to Red Hat Enterprise Linux 7 Laura Bailey Legal Notice Copyright © 2014 Red Hat, Inc. This document is licensed by Red Hat under the Creative Commons Attribution-ShareAlike 3.0 Unported License. If you distribute this document, or a modified version of it, you must provide attribution to Red Hat, Inc. and provide a link to the original. If the document is modified, all Red Hat trademarks must be removed. Red Hat, as the licensor of this document, waives the right to enforce, and agrees not to assert, Section 4d of CC-BY-SA to the fullest extent permitted by applicable law. Red Hat, Red Hat Enterprise Linux, the Shadowman logo, JBoss, MetaMatrix, Fedora, the Infinity Logo, and RHCE are trademarks of Red Hat, Inc., registered in the United States and other countries. Linux ® is the registered trademark of Linus Torvalds in the United States and other countries. Java ® is a registered trademark of Oracle and/or its affiliates. XFS ® is a trademark of Silicon Graphics International Corp. or its subsidiaries in the United States and/or other countries. MySQL ® is a registered trademark of MySQL AB in the United States, the European Union and other countries. Node.js ® is an official trademark of Joyent. Red Hat Software Collections is not formally related to or endorsed by the official Joyent Node.js open source or commercial project. The OpenStack ® Word Mark and OpenStack Logo are either registered trademarks/service marks or trademarks/service marks of the OpenStack Foundation, in the United States and other countries and are used with the OpenStack Foundation's permission. -

Porting Meego to LEON
#orting $eeGo to LE&' Master of Science Thesis ()A' *ERT&'A Chalmers University of Technology University of Gothenburg Department of Computer Science and Engineering Göteborg, Sweden, August !"" The Author grants to Chalmers University of Technology and University of Gothenburg the non-e-clusive right to publish the .or/ electronically and in a non-commercial purpose make it accessible on the (nternet0 The Author warrants that he1she is the author to the .ork, and warrants that the .or/ does not contain te-t, pictures or other material that violates copyright law0 The Author shall, when transferring the rights of the .or/ to a third party 2for e-ample a publisher or a company3, ac/nowledge the third party about this agreement0 (f the Author has signed a copyright agreement with a third party regarding the .ork, the Author warrants hereby that he1she has obtained any necessary permission from this third party to let Chalmers University of Technology and University of Gothenburg store the .or/ electronically and make it accessible on the (nternet0 #orting $eeGo to %E&' (van *ertona 4 (van *ertona, August !""0 E-aminer5 Arne Dahlberg Chalmers University of Technology University of Gothenburg Department of Computer Science and Engineering SE-6" 78 Göteborg Sweden Telephone 9 68 2!3:",;; "!!! Department of Computer Science and Engineering Göteborg, Sweden, August !"" POLITECNICO DI TORINO III Faculty of Information Engineering Master of Science in Computer Engineering Master Thesis Porting MeeGo To LEON Supervisor: Bartolomeo Montrucchio Candidate: Ivan Bertona August 2011 Abstract Portable multimedia devices are the flagship of a steadily growing market, from which the LEON/GRLIB hardware platform was excluded due to the lack of suitable software support. -

Chaussette Documentation Release 1.3.0
Chaussette Documentation Release 1.3.0 Tarek Ziade Apr 19, 2017 Contents 1 Usage 3 1.1 Running a plain WSGI application....................................3 1.2 Running a Django application......................................3 1.3 Running a Python Paste application...................................3 2 Using Chaussette in Circus 5 3 Using Chaussette in Supervisor7 4 Backends 9 5 Rationale and Design 11 6 Useful links 13 i ii Chaussette Documentation, Release 1.3.0 Chaussette is a WSGI server you can use to run your Python WSGI applications. The particularity of Chaussette is that it can either bind a socket on a port like any other server does or run against already opened sockets. That makes Chaussette the best companion to run a WSGI or Django stack under a process and socket manager, such as Circus or Supervisor. Contents 1 Chaussette Documentation, Release 1.3.0 2 Contents CHAPTER 1 Usage You can run a plain WSGI application, a Django application, or a Paste application. To get all options, just run chaussette –help. Running a plain WSGI application Chaussette provides a console script you can launch against a WSGI application, like any WSGI server out there: $ chaussette mypackage.myapp Application is <function myapp at 0x104d97668> Serving on localhost:8080 Using <class chaussette.backend._wsgiref.ChaussetteServer at 0x104e58d50> as a backend Running a Django application Chaussette allows you to run a Django project. You just need to provide the Python import path of the WSGI application, commonly located in the Django project’s wsgi.py file. For further information about how the wsgi. py file should look like see the Django documentation. -

An Introduction to HTTP Fingerprinting Table of Contents
An Introduction to HTTP fingerprinting Saumil Shah [email protected] 30th November, 2003 Table of Contents 1. Abstract 2. Theory of Fingerprinting 3. Banner Grabbing 4. Applications of HTTP Fingerprinting 5. Obfuscating the server banner string 6. Protocol Behaviour 6.1 HTTP Header field ordering 6.2 HTTP DELETE 6.3 Improper HTTP version response 6.3 Improper protocol response 6.5 Summary of test results 6.6 Choosing the right tests 7. Statistical and Fuzzy analysis 7.1 Assumptions 7.2 Terms and Definitions 7.3 Analysis Logic 8. httprint - the advanced HTTP fingerprinting engine 8.1 httprint signatures 8.2 httprint command line and GUI interfaces 8.3 Running httprint 8.4 The significance of confidence ratings 8.5 httprint Reports 8.6 Customising httprint 9. Trying to defeat HTTP Fingerprinting 10. Conclusion 11. References 1. Abstract HTTP Fingerprinting is a relatively new topic of discussion in the context of application security. One of the biggest challenges of maintaining a high level of network security is to have a complete and accurate inventory of networked assets. Web servers and web applications have now become a part of the scope of a network security assessment exercise. In this paper, we present techniques to identify various types of HTTP servers. We shall discuss some of the problems faced in inventorying HTTP servers and how we can overcome them. We shall also introduce and describe a tool, httprint, which is built using the concepts discussed in this paper. 2. Theory of Fingerprinting A fingerprint is defined as: 1. The impression of a fingertip on any surface; also: an ink impression of the lines upon the fingertip taken for the purpose of identification. -

Licensing Information User Manual Oracle Solaris 11.3 Last Updated September 2018
Licensing Information User Manual Oracle Solaris 11.3 Last Updated September 2018 Part No: E54836 September 2018 Licensing Information User Manual Oracle Solaris 11.3 Part No: E54836 Copyright © 2018, Oracle and/or its affiliates. All rights reserved. This software and related documentation are provided under a license agreement containing restrictions on use and disclosure and are protected by intellectual property laws. Except as expressly permitted in your license agreement or allowed by law, you may not use, copy, reproduce, translate, broadcast, modify, license, transmit, distribute, exhibit, perform, publish, or display any part, in any form, or by any means. Reverse engineering, disassembly, or decompilation of this software, unless required by law for interoperability, is prohibited. The information contained herein is subject to change without notice and is not warranted to be error-free. If you find any errors, please report them to us in writing. If this is software or related documentation that is delivered to the U.S. Government or anyone licensing it on behalf of the U.S. Government, then the following notice is applicable: U.S. GOVERNMENT END USERS: Oracle programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, delivered to U.S. Government end users are "commercial computer software" pursuant to the applicable Federal Acquisition Regulation and agency-specific supplemental regulations. As such, use, duplication, disclosure, modification, and adaptation of the programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, shall be subject to license terms and license restrictions applicable to the programs.