Morphological Variation of Two Howler Monkey Species and Their Genetically-Confirmed Hybrids
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Low Paternity Skew and the Influence of Maternal Kin in an Egalitarian
Low paternity skew and the influence of maternal kin in an egalitarian, patrilocal primate Karen B. Striera,1, Paulo B. Chavesb, Sérgio L. Mendesc, Valéria Fagundesc, and Anthony Di Fioreb,d aDepartment of Anthropology, University of Wisconsin, Madison, WI 53706; bDepartment of Anthropology and Center for the Study of Human Origins, New York University, New York, NY 10003; cDepartamento de Ciências Biológicas, Centro de Ciências Humanas e Naturais, Universidade Federal do Espírito Santo, Maruipe, Vitória, ES 29043-900, Brazil; and dDepartment of Anthropology, University of Texas at Austin, Austin, TX 78712 Contributed by Karen B. Strier, October 12, 2011 (sent for review September 11, 2011) Levels of reproductive skew vary in wild primates living in The fecal samples used for paternity analyses included the 22 multimale groups depending on the degree to which high-ranking infants born between 2005 and 2007 that survived to ≥2.08 y of males monopolize access to females. Still, the factors affecting age, their 21 mothers (representing diverse ages and histories) paternity in egalitarian societies remain unexplored. We combine (Table S1), and the 24 adult males that were possible sires of at unique behavioral, life history, and genetic data to evaluate the least one infant (Table S2). We considered a male to be a po- distribution of paternity in the northern muriqui (Brachyteles tential sire for an infant if he was >5 y and was known to have hypoxanthus), a species known for its affiliative, nonhierarchical completed a copulation before the infant’s estimated conception relationships. We genotyped 67 individuals (22 infants born over a date (birth date minus mean 216.4-d gestation) (16). -

Diets of Howler Monkeys
Chapter 2 Diets of Howler Monkeys Pedro Américo D. Dias and Ariadna Rangel-Negrín Abstract Based on a bibliographical review, we examined the diets of howler mon- keys to compile a comprehensive overview of their food resources and document dietary diversity. Additionally, we analyzed the effects of rainfall, group size, and forest size on dietary variation. Howlers eat nearly all available plant parts in their habitats. Time dedicated to the consumption of different food types varies among species and populations, such that feeding behavior can range from high folivory to high frugivory. Overall, howlers were found to use at least 1,165 plant species, belonging to 479 genera and 111 families as food sources. Similarity in the use of plant taxa as food sources (assessed with the Jaccard index) is higher within than between howler species, although variation in similarity is higher within species. Rainfall patterns, group size, and forest size affect several dimensions of the dietary habits of howlers, such that, for instance, the degree of frugivory increases with increased rainfall and habitat size, but decreases with increasing group size in groups that live in more productive habitats. Moreover, the range of variation in dietary habits correlates positively with variation in rainfall, suggesting that some howler species are habitat generalists and have more variable diets, whereas others are habi- tat specialists and tend to concentrate their diets on certain plant parts. Our results highlight the high degree of dietary fl exibility demonstrated by the genus Alouatta and provide new insights for future research on howler foraging strategies. Resumen Con base en una revisión bibliográfi ca, examinamos las dietas de los monos aulladores para describir exhaustivamente sus recursos alimenticios y la diversidad de su dieta. -
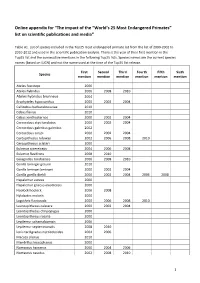
Online Appendix for “The Impact of the “World's 25 Most Endangered
Online appendix for “The impact of the “World’s 25 Most Endangered Primates” list on scientific publications and media” Table A1. List of species included in the Top25 most endangered primate list from the list of 2000-2002 to 2010-2012 and used in the scientific publication analysis. There is the year of their first mention in the Top25 list and the consecutive mentions in the following Top25 lists. Species names are the current species names (based on IUCN) and not the name used at the time of the Top25 list release. First Second Third Fourth Fifth Sixth Species mention mention mention mention mention mention Ateles fusciceps 2006 Ateles hybridus 2006 2008 2010 Ateles hybridus brunneus 2004 Brachyteles hypoxanthus 2000 2002 2004 Callicebus barbarabrownae 2010 Cebus flavius 2010 Cebus xanthosternos 2000 2002 2004 Cercocebus atys lunulatus 2000 2002 2004 Cercocebus galeritus galeritus 2002 Cercocebus sanjei 2000 2002 2004 Cercopithecus roloway 2002 2006 2008 2010 Cercopithecus sclateri 2000 Eulemur cinereiceps 2004 2006 2008 Eulemur flavifrons 2008 2010 Galagoides rondoensis 2006 2008 2010 Gorilla beringei graueri 2010 Gorilla beringei beringei 2000 2002 2004 Gorilla gorilla diehli 2000 2002 2004 2006 2008 Hapalemur aureus 2000 Hapalemur griseus alaotrensis 2000 Hoolock hoolock 2006 2008 Hylobates moloch 2000 Lagothrix flavicauda 2000 2006 2008 2010 Leontopithecus caissara 2000 2002 2004 Leontopithecus chrysopygus 2000 Leontopithecus rosalia 2000 Lepilemur sahamalazensis 2006 Lepilemur septentrionalis 2008 2010 Loris tardigradus nycticeboides -

World's Most Endangered Primates
Primates in Peril The World’s 25 Most Endangered Primates 2016–2018 Edited by Christoph Schwitzer, Russell A. Mittermeier, Anthony B. Rylands, Federica Chiozza, Elizabeth A. Williamson, Elizabeth J. Macfie, Janette Wallis and Alison Cotton Illustrations by Stephen D. Nash IUCN SSC Primate Specialist Group (PSG) International Primatological Society (IPS) Conservation International (CI) Bristol Zoological Society (BZS) Published by: IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), Bristol Zoological Society (BZS) Copyright: ©2017 Conservation International All rights reserved. No part of this report may be reproduced in any form or by any means without permission in writing from the publisher. Inquiries to the publisher should be directed to the following address: Russell A. Mittermeier, Chair, IUCN SSC Primate Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, VA 22202, USA. Citation (report): Schwitzer, C., Mittermeier, R.A., Rylands, A.B., Chiozza, F., Williamson, E.A., Macfie, E.J., Wallis, J. and Cotton, A. (eds.). 2017. Primates in Peril: The World’s 25 Most Endangered Primates 2016–2018. IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society, Arlington, VA. 99 pp. Citation (species): Salmona, J., Patel, E.R., Chikhi, L. and Banks, M.A. 2017. Propithecus perrieri (Lavauden, 1931). In: C. Schwitzer, R.A. Mittermeier, A.B. Rylands, F. Chiozza, E.A. Williamson, E.J. Macfie, J. Wallis and A. Cotton (eds.), Primates in Peril: The World’s 25 Most Endangered Primates 2016–2018, pp. 40-43. IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society, Arlington, VA. -

Vânia Luciane Alves Garcia
Neotropical Primates 13(Suppl.), December 2005 79 SURVEY AND STATUS OF THE MURIQUIS BRACHYTELES ARACHNOIDES IN THE SERRA DOS ÓRGÃOS NATIONAL PARK, RIO DE JANEIRO Vânia Luciane Alves Garcia Seção de Mastozoologia, Departamento de Vertebrados, Museu Nacional, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil, e-mail: <[email protected]> Abstract Th e Serra dos Órgãos National Park protects 11,800 ha of Atlantic forest in the state of Rio de Janeiro (22°30'S, 43°06'W; 300 to 2,263 m above sea level). Vegetation types include montane dense evergreeen forest up to altitudes of 1,800 m; cloud forest from 1,800 to 2,000 m; and high altitude grassland above 2,000 m. Th is paper reports on surveys carried out in 10 localities in the park specifi cally to obtain a minimum estimate of the population of northern muriquis (Brachyteles arachnoides). Muriquis were sighted 26 times at altitudes ranging from 800 to 1,500 m. It is possible that they belonged to four groups in which case the minimum number of individuals recorded would be 56. If in fact the sightings were of just two groups, the minimum number of individuals would be 32. Black-horned capuchin (Cebus nigritus) and brown howler mon- keys (Alouatta guariba) were also recorded for the park. Th e buff y-tufted-ear marmoset (Callithrix aurita) was not seen. Key Words – primates, muriqui, Brachyteles, Serra dos Órgãos, Atlantic forest, Brazil Introduction (22°30'S, 43°06'W) (Fig. 1). A number of rivers supplying water to urban centers in the lowlands have their sources in Th ere is now some quite substantial information on the these mountains, including the rios Paquequer, Soberbo, populations of the northern muriqui in Minas Gerais Jacó, Bananal, Bonfi m and Santo Aleixo. -

Neotropical Primates 20(1), June 2012
ISSN 1413-4703 NEOTROPICAL PRIMATES A Journal of the Neotropical Section of the IUCN/SSC Primate Specialist Group Volume 20 Number 1 June 2013 Editors Erwin Palacios Liliana Cortés-Ortiz Júlio César Bicca-Marques Eckhard Heymann Jessica Lynch Alfaro Anita Stone News and Book Reviews Brenda Solórzano Ernesto Rodríguez-Luna PSG Chairman Russell A. Mittermeier PSG Deputy Chairman Anthony B. Rylands Neotropical Primates A Journal of the Neotropical Section of the IUCN/SSC Primate Specialist Group Conservation International 2011 Crystal Drive, Suite 500, Arlington, VA 22202, USA ISSN 1413-4703 Abbreviation: Neotrop. Primates Editors Erwin Palacios, Conservación Internacional Colombia, Bogotá DC, Colombia Liliana Cortés Ortiz, Museum of Zoology, University of Michigan, Ann Arbor, MI, USA Júlio César Bicca-Marques, Pontifícia Universidade Católica do Rio Grande do Sul, Porto Alegre, Brasil Eckhard Heymann, Deutsches Primatenzentrum, Göttingen, Germany Jessica Lynch Alfaro, Institute for Society and Genetics, University of California-Los Angeles, Los Angeles, CA, USA Anita Stone, Department of Biology, Eastern Michigan University, Ypsilanti, MI, USA News and Books Reviews Brenda Solórzano, Instituto de Neuroetología, Universidad Veracruzana, Xalapa, México Ernesto Rodríguez-Luna, Instituto de Neuroetología, Universidad Veracruzana, Xalapa, México Founding Editors Anthony B. Rylands, Center for Applied Biodiversity Science Conservation International, Arlington VA, USA Ernesto Rodríguez-Luna, Instituto de Neuroetología, Universidad Veracruzana, Xalapa, México Editorial Board Bruna Bezerra, University of Louisville, Louisville, KY, USA Hannah M. Buchanan-Smith, University of Stirling, Stirling, Scotland, UK Adelmar F. Coimbra-Filho, Academia Brasileira de Ciências, Rio de Janeiro, Brazil Carolyn M. Crockett, Regional Primate Research Center, University of Washington, Seattle, WA, USA Stephen F. Ferrari, Universidade Federal do Sergipe, Aracajú, Brazil Russell A. -

Central American Spider Monkey Ateles Geoffroyi Kuhl, 1820: Mexico, Guatemala, Nicaragua, Honduras, El Salvador
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/321428630 Central American Spider Monkey Ateles geoffroyi Kuhl, 1820: Mexico, Guatemala, Nicaragua, Honduras, El Salvador... Chapter · December 2017 CITATIONS READS 0 18 7 authors, including: Pedro Guillermo Mendez-Carvajal Gilberto Pozo-Montuy Fundacion Pro-Conservacion de los Primates… Conservación de la Biodiversidad del Usuma… 13 PUBLICATIONS 24 CITATIONS 32 PUBLICATIONS 202 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Connectivity of priority sites for primate conservation in the Zoque Rainforest Complex in Southeastern Mexico View project Regional Monitoring System: communitarian participation in surveying Mexican primate’s population (Ateles and Alouatta) View project All content following this page was uploaded by Gilberto Pozo-Montuy on 01 December 2017. The user has requested enhancement of the downloaded file. Primates in Peril The World’s 25 Most Endangered Primates 2016–2018 Edited by Christoph Schwitzer, Russell A. Mittermeier, Anthony B. Rylands, Federica Chiozza, Elizabeth A. Williamson, Elizabeth J. Macfie, Janette Wallis and Alison Cotton Illustrations by Stephen D. Nash IUCN SSC Primate Specialist Group (PSG) International Primatological Society (IPS) Conservation International (CI) Bristol Zoological Society (BZS) Published by: IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), Bristol Zoological Society (BZS) Copyright: ©2017 Conservation International All rights reserved. No part of this report may be reproduced in any form or by any means without permission in writing from the publisher. Inquiries to the publisher should be directed to the following address: Russell A. -

Impact of Intrasexual Selection on Sexual Dimorphism and Testes Size in the Mexican Howler Monkeys Alouatta Palliata and A
AMERICAN JOURNAL OF PHYSICAL ANTHROPOLOGY 146:179–187 (2011) Impact of Intrasexual Selection on Sexual Dimorphism and Testes Size in the Mexican Howler Monkeys Alouatta palliata and A. pigra Mary Kelaita,1* Pedro Ame´ rico D. Dias,2 Ma. del Socorro Aguilar-Cucurachi,2 Domingo Canales-Espinosa,2 and Liliana Corte´ s-Ortiz3 1Department of Anthropology, University of Michigan, Ann Arbor, MI 48109-1107 2Instituto de Neuroetologı´a, Universidad Veracruzana, Xalapa, Mexico 3Museum of Zoology and Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109-1079 KEY WORDS body size dimorphism; canine dimorphism; sperm competition ABSTRACT One of the goals of physical anthropology body mass and length, and dental data were obtained and primatology is to understand how primate social sys- from casts from wild individuals and from museum speci- tems influence the evolution of sexually selected traits. mens. Although A. pigra individuals are larger than their Howler monkeys provide a good model for studying sex- A. palliata counterparts, we find that both species exhibit ual selection due to differences in social systems between similar levels of sexual dimorphism for all of the variables related species. Here, we examine data from the sister considered. Testicular volume results indicate that A. pal- howler monkey species Alouatta palliata and A. pigra liata male testes are on average twice as large as those of inhabiting southeastern Mexico and northern Guatemala. A. pigra males, suggesting more intense sperm competi- We use a resampling approach to analyze differences in tion in the former species. Our study shows that A. pigra sexual dimorphism of body and canine size. -

Conservation of the Brown Howler Monkey in South-East Brazil
ORYX VOL 28 NO 1 JANUARY 1994 Conservation of the brown howler monkey in south-east Brazil Adriano Garcia Chiarello and Mauro Galetti The brown howler monkey Alouatta fusca once had a wide geographical distribution throughout a large part of the Atlantic forest in Brazil. Today only 5 per cent of these forests remain and the species is endangered. Howler monkeys can thrive in small forest fragments but they are more vulnerable to hunting, disease, and predation in these habitats than in undisturbed forests. Brown howler monkeys are important seed dispersers of several plant species, particularly in isolated forest fragments where specialized frugivores are absent. In protected areas without large predators howlers can reach high densities and the management of these populations is necessary to avoid inbreeding. Introduction on free-ranging brown howler monkeys started only recently (Mendes, 1989; Prates et The Atlantic forest of Brazil originally ex- al., 1990; Chiarello, 1992; Galetti, 1992). The tended from the state of Rio Grande do Norte species occurs in southern Bahia, Espirito to the southernmost borders of the country, Santo, eastern Minas Gerais, Rio de Janeiro, into the northern Argentinian province of Sao Paulo, Parana, Santa Catarina and north- Missiones and parts of eastern Paraguay. This ern Rio Grande do Sul in Brazil, as well as in forest once covered more than 1 million sq the extreme north-east of Argentina (Figure 1). km, making it the third most extensive veg- Ihering (1914) recognized two subspecies, etation type in Brazil after the Amazonian for- the northern brown howler A. fusca fusca from est and the cerrado (Mittermeier et al., 1989). -

Neotropical Primates 17(1), June 2010
ISSN 1413-4703 NEOTROPICAL PRIMATES A Journal of the Neotropical Section of the IUCN/SSC Primate Specialist Group Volume 17 Number 2 December 2010 Editors Erwin Palacios Liliana Cortés-Ortiz Júlio César Bicca-Marques Eckhard Heymann Jessica Lynch Alfaro Liza Veiga News and Book Reviews Brenda Solórzano Ernesto Rodríguez-Luna PSG Chairman Russell A. Mittermeier PSG Deputy Chairman SPECIES SURVIVAL Anthony B. Rylands COMMISSION Neotropical Primates A Journal of the Neotropical Section of the IUCN/SSC Primate Specialist Group Conservation International 2011 Crystal Drive, Suite 500, Arlington, VA 22202, USA ISSN 1413-4703 Abbreviation: Neotrop. Primates Editors Erwin Palacios, Conservación Internacional Colombia, Bogotá DC, Colombia Liliana Cortés Ortiz, Museum of Zoology, University of Michigan, Ann Arbor, MI, USA Júlio César Bicca-Marques, Pontifícia Universidade Católica do Rio Grande do Sul, Porto Alegre, Brasil Eckhard Heymann, Deutsches Primatenzentrum, Göttingen, Germany Jessica Lynch Alfaro, Washington State University, Pullman, WA, USA Liza Veiga, Museu Paraense Emílio Goeldi, Belém, Brazil News and Books Reviews Brenda Solórzano, Instituto de Neuroetología, Universidad Veracruzana, Xalapa, México Ernesto Rodríguez-Luna, Instituto de Neuroetología, Universidad Veracruzana, Xalapa, México Founding Editors Anthony B. Rylands, Center for Applied Biodiversity Science Conservation International, Arlington VA, USA Ernesto Rodríguez-Luna, Instituto de Neuroetología, Universidad Veracruzana, Xalapa, México Editorial Board Bruna Bezerra, University of Louisville, Louisville, KY, USA Hannah M. Buchanan-Smith, University of Stirling, Stirling, Scotland, UK Adelmar F. Coimbra-Filho, Academia Brasileira de Ciências, Rio de Janeiro, Brazil Carolyn M. Crockett, Regional Primate Research Center, University of Washington, Seattle, WA, USA Stephen F. Ferrari, Universidade Federal do Sergipe, Aracajú, Brazil Russell A. -

Laboratory Primate Newsletter
LABORATORY PRIMATE NEWSLETTER Vol. 49, No. 4 October 2010 JUDITH E. SCHRIER, EDITOR JAMES S. HARPER, GORDON J. HANKINSON AND LARRY HULSEBOS, ASSOCIATE EDITORS MORRIS L. POVAR AND JASON MACHAN, CONSULTING EDITORS ELVA MATHIESEN, ASSISTANT EDITOR ALLAN M. SCHRIER, FOUNDING EDITOR, 1962-1987 Published Quarterly by the Schrier Research Laboratory Psychology Department, Brown University Providence, Rhode Island ISSN 0023-6861 POLICY STATEMENT The Laboratory Primate Newsletter provides a central source of information about nonhuman primates and related matters to scientists who use these animals in their research and those whose work supports such research. The Newsletter (1) provides information on care and breeding of nonhuman primates for laboratory research, (2) disseminates general information and news about the world of primate research (such as announcements of meetings, research projects, sources of information, nomenclature changes), (3) helps meet the special research needs of individual investigators by publishing requests for research material or for information related to specific research problems, and (4) serves the cause of conservation of nonhuman primates by publishing information on that topic. As a rule, research articles or summaries accepted for the Newsletter have some practical implications or provide general information likely to be of interest to investigators in a variety of areas of primate research. However, special consideration will be given to articles containing data on primates not conveniently publishable elsewhere. General descriptions of current research projects on primates will also be welcome. The Newsletter appears quarterly and is intended primarily for persons doing research with nonhuman primates. Back issues may be purchased for $10.00 each. We are no longer printing paper issues, except those we will send to subscribers who have paid in advance. -

Alouatta Pigra) John F
www.nature.com/scientificreports OPEN Spatial aggregation of fruits explains food selection in a neotropical primate (Alouatta pigra) John F. Aristizabal 1, Simoneta Negrete-Yankelevich 2*, Rogelio Macías-Ordóñez 3, Colin A. Chapman 4,5,6 & Juan C. Serio-Silva 1 The availability and spatial distribution of food resources afect animal behavior and survival. Black howler monkeys (Alouatta pigra) have a foraging strategy to balance their nutrient intake that involves mixing their consumption of leaves and fruits. The spatial aggregation of food items should impact this strategy, but how it does so is largely unknown. We quantifed how leaf and fruit intake combined (here termed food set selection) was spatially aggregated in patches and how food aggregation varied across seasons. Using variograms we estimated patch diameter and with Generalized Least Square models determined the efect of food spatial aggregation on food selection. Only fruits were structured in patches in the season of highest availability (dry-season). The patches of food set selection had a diameter between 6.9 and 14 m and were explained by those of mature fruit availability which were between 18 and 19 m in diameter. Our results suggest that the spatial pattern of food selection is infuenced by patches of large fruit-bearing trees, not by particular species. Fruit also occur along spatial gradients, but these do not explain food selection, suggesting that howlers maximize food intake in response to local aggregation of fruit that are limiting during certain seasons. We demonstrate how the independent spatial modelling of resources and behavior enables the defnition of patches and testing their spatial relationship.