1
Supplemental Table S6. Genes up- and down-regulated by CCCP
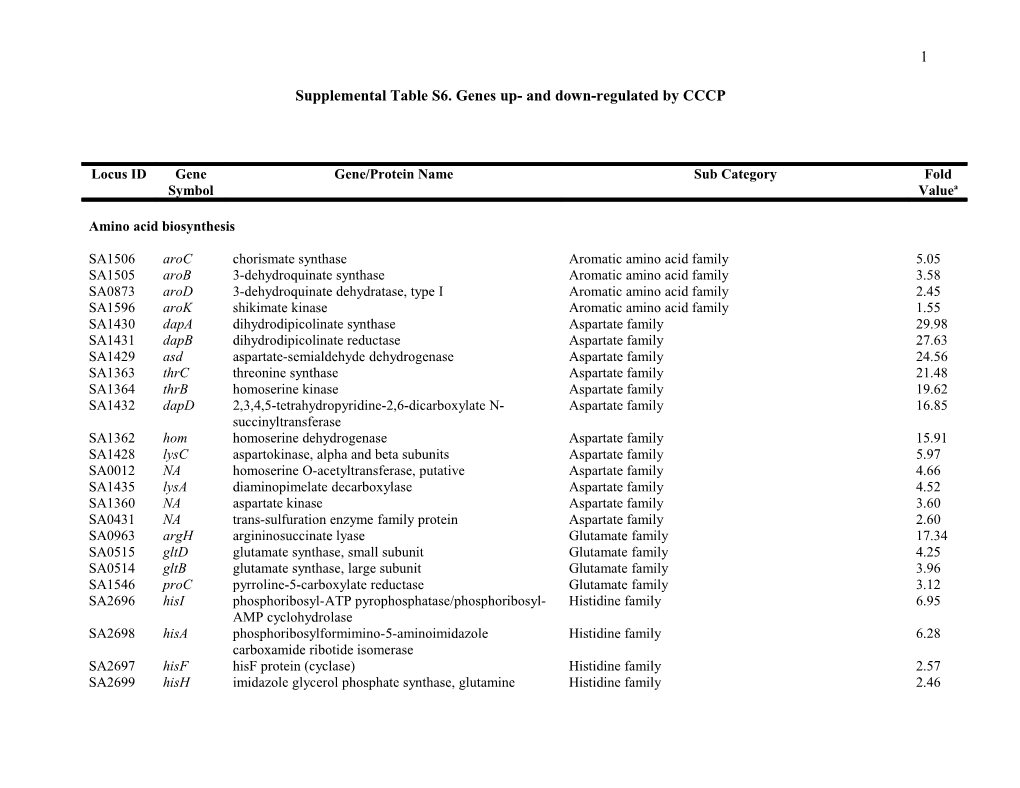
Locus ID Gene Gene/Protein Name Sub Category Fold Symbol Valuea
Amino acid biosynthesis
SA1506 aroC chorismate synthase Aromatic amino acid family 5.05 SA1505 aroB 3-dehydroquinate synthase Aromatic amino acid family 3.58 SA0873 aroD 3-dehydroquinate dehydratase, type I Aromatic amino acid family 2.45 SA1596 aroK shikimate kinase Aromatic amino acid family 1.55 SA1430 dapA dihydrodipicolinate synthase Aspartate family 29.98 SA1431 dapB dihydrodipicolinate reductase Aspartate family 27.63 SA1429 asd aspartate-semialdehyde dehydrogenase Aspartate family 24.56 SA1363 thrC threonine synthase Aspartate family 21.48 SA1364 thrB homoserine kinase Aspartate family 19.62 SA1432 dapD 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N- Aspartate family 16.85 succinyltransferase SA1362 hom homoserine dehydrogenase Aspartate family 15.91 SA1428 lysC aspartokinase, alpha and beta subunits Aspartate family 5.97 SA0012 NA homoserine O-acetyltransferase, putative Aspartate family 4.66 SA1435 lysA diaminopimelate decarboxylase Aspartate family 4.52 SA1360 NA aspartate kinase Aspartate family 3.60 SA0431 NA trans-sulfuration enzyme family protein Aspartate family 2.60 SA0963 argH argininosuccinate lyase Glutamate family 17.34 SA0515 gltD glutamate synthase, small subunit Glutamate family 4.25 SA0514 gltB glutamate synthase, large subunit Glutamate family 3.96 SA1546 proC pyrroline-5-carboxylate reductase Glutamate family 3.12 SA2696 hisI phosphoribosyl-ATP pyrophosphatase/phosphoribosyl- Histidine family 6.95 AMP cyclohydrolase SA2698 hisA phosphoribosylformimino-5-aminoimidazole Histidine family 6.28 carboxamide ribotide isomerase SA2697 hisF hisF protein (cyclase) Histidine family 2.57 SA2699 hisH imidazole glycerol phosphate synthase, glutamine Histidine family 2.46 2
amidotransferase subunit SA2045 ilvC ketol-acid reductoisomerase Pyruvate family 12.07 SA2043 ilvB acetolactate synthase, large subunit, biosynthetic type Pyruvate family 9.98 SA0600 ilvE branched-chain amino acid aminotransferase Pyruvate family 5.52 SA2047 leuB 3-isopropylmalate dehydrogenase Pyruvate family 2.27 SA2046 leuA 2-isopropylmalate synthase Pyruvate family 1.84 SA1773 serA D-3-phosphoglycerate dehydrogenase Serine family 3.65 SA2105 glyA serine hydroxymethyltransferase Serine family 2.13
Purines, pyrimidines, nucleosides, and nucleotides
SA0130 NA 5 nucleotidase family protein Other 2.39 SA0123 deoC1 deoxyribose-phosphate aldolase Other 2.34 SA1031 NA 5 nucleotidase family protein Other 2.14 SA0124 deoB phosphopentomutase Other 1.70 SA1625 cdd cytidine deaminase Salvage of nucleosides and nucleotides 2.20 SA2104 upp uracil phosphoribosyltransferase Salvage of nucleosides and nucleotides 1.98
Fatty acid and phospholipid metabolism
SA1351 cls1 cardiolipin synthetase Biosynthesis 5.12 SA2061 acpS holo-(acyl-carrier-protein) synthase Biosynthesis 3.16 SA2482 fabG2 3-oxoacyl-(acyl carrier protein) reductase, authentic Biosynthesis 2.00 point mutation SA1302 pgsA CDP-diacylglycerol--glycerol-3-phosphate 3- Biosynthesis 1.79 phosphatidyltransferase SA2651 NA tributyrin esterase EstA, putative Degradation 5.29 SA1814 NA lysophospholipase, putative Degradation 1.85 SA1439 NA acylphosphatase Other 3.79
Biosynthesis of cofactors, prosthetic groups, and carriers
SA2105 glyA serine hydroxymethyltransferase Folic acid 2.13 SA2641 gpxA2 glutathione peroxidase Glutathione and analogs 3.45 SA1325 gpxA1 glutathione peroxidase Glutathione and analogs 3.17 SA1889 hemE uroporphyrinogen decarboxylase Heme, porphyrin, and cobalamin 2.19 SA1922 hemL2 glutamate-1-semialdehyde-2,1-aminomutase Heme, porphyrin, and cobalamin 1.72 3
SA2264 moaE molybdenum cofactor biosynthesis protein E Molybdopterin 3.94 SA2268 moaB molybdenum cofactor biosynthesis protein B Molybdopterin 3.64 SA2269 NA molybdopterin biosynthesis MoeB protein, putative Molybdopterin 1.78 SA2261 moaA molybdenum cofactor biosynthesis protein A Molybdopterin 1.69 SA2267 moaC molybdenum cofactor biosynthesis protein C Molybdopterin 1.69 SA0236 NA 4-diphosphocytidyl-2C-methyl-D-erythritol synthase, Other 8.44 putative SA0918 sufB FeS assembly protein SufB Other 1.97 SA0915 sufD FeS assembly protein SufD Other 1.50 SA2615 panB 3-methyl-2-oxobutanoate hydroxymethyltransferase Pantothenate and coenzyme A 4.89 SA2614 panC pantoate--beta-alanine ligase Pantothenate and coenzyme A 3.43 SA1223 coaBC phosphopantothenoylcysteine Pantothenate and coenzyme A 2.94 decarboxylase/phosphopantothenate--cysteine ligase SA2448 NA 2-dehydropantoate 2-reductase, putative Pantothenate and coenzyme A 2.13 SA2616 NA 2-dehydropantoate 2-reductase PanE, putative Pantothenate and coenzyme A 2.09 SA1974 nadE NH(3)-dependent NAD+ synthetase Pyridine nucleotides 2.71 SA1291 ribF riboflavin biosynthesis protein RibF Riboflavin, FMN, and FAD 2.56
Central intermediary metabolism
SA0616 nagB glucosamine-6-phosphate isomerase Amino sugars 2.40 SA2280 ureA urease, gamma subunit Nitrogen metabolism 6.49 SA2283 ureE urease accessory protein UreE Nitrogen metabolism 5.40 SA2282 ureC urease, alpha subunit Nitrogen metabolism 4.93 SA2281 ureB urease, beta subunit Nitrogen metabolism 3.50 SA2285 ureG urease accessory protein UreG Nitrogen metabolism 3.45 SA2286 ureD urease accessory protein UreD Nitrogen metabolism 2.58 SA2284 ureF urease accessory protein UreF Nitrogen metabolism 2.30 SA1920 NA D-isomer specific 2-hydroxyacid dehydrogenase family Other 4.01 protein SA2570 NA galactoside O-acetyltransferase Other 2.28 SA2574 NA D-isomer specific 2-hydroxyacid dehydrogenase family Other 2.19 protein SA0634 pta phosphate acetyltransferase Other 2.03 SA2577 crtM dehydrosqualene synthase Other 1.62 SA1982 ppaC manganese-dependent inorganic pyrophosphatase Phosphorus compounds 2.44 4
Energy metabolism
SA0973 NA fumarylacetoacetate hydrolase family protein Amino acids and amines 2.16 SA2656 arcB2 ornithine carbamoyltransferase Amino acids and amines 2.13 SA0877 gcvH glycine cleavage system H protein Amino acids and amines 1.59 SA2273 fdhD formate dehydrogenase accessory protein FdhD Anaerobic 2.26 SA0162 NA formate dehydrogenase, NAD-dependent Anaerobic 1.91 SA2534 frp NAD(P)H-flavin oxidoreductase Electron transport 4.91 SA0453 NA NAD(P)H-flavin oxidoreductase, putative Electron transport 3.66 SA0829 trxB thioredoxin reductase Electron transport 3.09 SA0494 nuoF NADH dehydrogenase I, F subunit Electron transport 2.72 SA0875 NA thioredoxin, putative Electron transport 2.64 SA1794 NA thioredoxin, putative Electron transport 2.34 SA0190 NA NAD(P)H dehydrogenase (quinone), putative Electron transport 2.30 SA2273 fdhD formate dehydrogenase accessory protein FdhD Electron transport 2.26 SA0941 NA NADH dehydrogenase, putative, authentic frameshift Electron transport 1.88 SA1155 trxA thioredoxin Electron transport 1.79 SA2178 NA alcohol dehydrogenase, zinc-containing Fermentation 9.97 SA2114 NA aldehyde dehydrogenase Fermentation 4.35 SA2177 NA alcohol dehydrogenase, zinc-containing Fermentation 3.90 SA0241 NA alcohol dehydrogenase, zinc-containing Fermentation 2.19 SA1984 aldA2 aldehyde dehydrogenase Fermentation 2.14 SA0634 pta phosphate acetyltransferase Fermentation 2.03 SA0839 pgk phosphoglycerate kinase Glycolysis/gluconeogenesis 8.35 SA0838 gapA1 glyceraldehyde 3-phosphate dehydrogenase Glycolysis/gluconeogenesis 6.50 SA0841 pgm phosphoglycerate mutase, 2,3-bisphosphoglycerate- Glycolysis/gluconeogenesis 4.70 independent SA2415 gpm phosphoglycerate mutase Glycolysis/gluconeogenesis 4.23 SA0840 tpiA triosephosphate isomerase Glycolysis/gluconeogenesis 3.94 SA2622 fdaB fructose-bisphosphate aldolase, class I Glycolysis/gluconeogenesis 3.52 SA1746 pfkA 6-phosphofructokinase Glycolysis/gluconeogenesis 3.22 SA1123 pyc pyruvate carboxylase Glycolysis/gluconeogenesis 2.48 SA0842 eno enolase Glycolysis/gluconeogenesis 2.26 SA0966 pgi glucose-6-phosphate isomerase Glycolysis/gluconeogenesis 2.06 SA1745 pyk pyruvate kinase Glycolysis/gluconeogenesis 2.05 SA2624 NA acetyl-CoA synthetase, putative Other 6.72 SA0805 NA glycerate kinase family protein Other 2.80 5
SA0123 deoC1 deoxyribose-phosphate aldolase Other 2.34 SA0973 NA fumarylacetoacetate hydrolase family protein Other 2.16 SA2553 NA pyruvate oxidase Other 1.54 SA2435 NA glycerate kinase Other 1.52 SA1831 tal transaldolase Pentose phosphate pathway 6.87 SA1549 zwf glucose-6-phosphate 1-dehydrogenase Pentose phosphate pathway 2.83 SA1377 tkt transketolase Pentose phosphate pathway 2.28 SA2332 galM aldose 1-epimerase Sugars 2.19 SA0617 NA hexulose-6-phosphate synthase, putative Sugars 1.99 SA0235 NA hexitol dehydrogenase Sugars 1.62
Transport and binding proteins
SA2619 NA amino acid permease Amino acids, peptides and amines 12.60 SA2453 NA amino acid ABC transporter, ATP-binding protein Amino acids, peptides and amines 7.34 SA0994 oppF oligopeptide ABC transporter, ATP-binding protein Amino acids, peptides and amines 6.81 SA2451 NA amino acid ABC transporter, amino acid-binding protein Amino acids, peptides and amines 6.75 SA1915 NA amino acid ABC transporter, ATP-binding protein Amino acids, peptides and amines 6.54 SA1916 NA amino acid ABC transporter, permease/substrate- Amino acids, peptides and amines 5.38 binding protein SA2450 NA amino acid ABC transporter, permease protein Amino acids, peptides and amines 5.17 SA0995 NA oligopeptide ABC transporter, oligopeptide-binding Amino acids, peptides and amines 5.11 protein SA0781 NA osmoprotectant ABC transporter, ATP-binding protein, Amino acids, peptides and amines 4.80 putative SA2452 NA amino acid ABC transporter, permease protein Amino acids, peptides and amines 4.63 SA0991 oppB oligopeptide ABC transporter, permease protein Amino acids, peptides and amines 3.07 SA1728 NA amino acid permease Amino acids, peptides and amines 2.95 SA1963 NA proline permease Amino acids, peptides and amines 2.18 SA0993 oppD oligopeptide ABC transporter, ATP-binding protein Amino acids, peptides and amines 2.10 SA2458 NA amino acid permease Amino acids, peptides and amines 1.76 SA2272 modA molybdenum ABC transporter, molybdenum-binding Anions 2.39 protein ModA SA1424 NA phosphate ABC transporter, phosphate-binding protein Anions 2.01 SA2270 modC molybdenum ABC transporter, ATP-binding protein Anions 1.65 ModC SA1457 NA PTS system, IIA component Carbohydrates, organic alcohols, and acids 3.37 6
SA0224 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids 2.41 SA0136 cap5A capsular polysaccharide biosynthesis protein Cap5A Carbohydrates, organic alcohols, and acids 1.90 SA2181 lacE PTS system, lactose-specific IIBC components Carbohydrates, organic alcohols, and acids 1.64 SA2068 kdpA potassium-transporting ATPase, A subunit Cations and iron carrying compounds 13.40 SA2067 kdpB potassium-transporting P-type ATPase, B subunit Cations and iron carrying compounds 11.10 SA2066 kdpC potassium-transporting ATPase, C subunit Cations and iron carrying compounds 6.76 SA1114 NA Mn2+/Fe2+ transporter, NRAMP family Cations and iron carrying compounds 4.60 SA2572 NA copper-translocating P-type ATPase Cations and iron carrying compounds 3.79 SA1919 NA transcriptional regulator, Fur family Cations and iron carrying compounds 3.24 SA0687 NA Na+/H+ antiporter, putative Cations and iron carrying compounds 2.85 SA0704 NA iron compound ABC transporter, ATP-binding protein Cations and iron carrying compounds 2.53 SA2167 NA iron compound ABC transporter, iron compound- Cations and iron carrying compounds 2.29 binding protein SA1013 mgtE magnesium transporter Cations and iron carrying compounds 1.92 SA1541 NA transcriptional regulator, Fur family Cations and iron carrying compounds 1.88 SA0105 NA siderophore biosynthesis protein, IucC family Cations and iron carrying compounds 1.53 SA1007 NA protozoan/cyanobacterial globin family protein Other 3.84 SA0086 NA drug transporter, putative Other 3.74 SA2449 NA drug transporter, putative Other 3.05 SA0163 NA drug transporter, putative Other 2.48 SA2413 NA drug resistance transporter, EmrB/QacA subfamily Other 1.85 SA0882 NA ABC transporter, ATP-binding protein Unknown substrate 16.30 SA0884 NA ABC transporter, substrate-binding protein Unknown substrate 11.09 SA0883 NA ABC transporter, permease protein Unknown substrate 11.04 SA0700 abcA ABC transporter, ATP-binding/permease protein Unknown substrate 3.65 SA0506 NA ABC transporter, substrate-binding protein Unknown substrate 2.91 SA0504 NA ABC transporter, ATP-binding protein Unknown substrate 2.76 SA2708 NA ABC transporter, ATP-binding protein Unknown substrate 2.71 SA2643 NA ABC transporter, permease protein Unknown substrate 2.38 SA2279 NA transporter, putative Unknown substrate 2.30 SA2144 NA ABC transporter, ATP-binding protein Unknown substrate 2.03 SA0744 NA ABC transporter, ATP-binding protein, MsbA family Unknown substrate 1.67 SA2483 NA transporter, putative Unknown substrate 1.53
DNA metabolism
SA1479 NA 5-3 exonuclease, putative Degradation of DNA 1.75 7
SA2562 ogt methylated-DNA--protein-cysteine methyltransferase DNA replication, recombination, and repair 7.83 SA0970 rexB exonuclease RexB DNA replication, recombination, and repair 5.16 SA1732 NA replication initiation and membrane attachment protein DNA replication, recombination, and repair 4.24 SA1955 dinP DNA-damage-inducible protein P DNA replication, recombination, and repair 4.10 SA1493 NA DNA replication protein DnaD, putative DNA replication, recombination, and repair 3.94 SA1736 fpg formamidopyrimidine-DNA glycosylase DNA replication, recombination, and repair 3.88 SA1737 polA DNA polymerase I DNA replication, recombination, and repair 2.92 SA1540 xerD site-specific recombinase XerD DNA replication, recombination, and repair 2.86 SA0971 rexA exonuclease RexA DNA replication, recombination, and repair 2.77 SA1224 priA primosomal protein N` DNA replication, recombination, and repair 2.45 SA1269 xerC integrase/recombinase DNA replication, recombination, and repair 2.33 SA1241 recG ATP-dependent DNA helicase RecG DNA replication, recombination, and repair 2.18 SA1623 NA DNA repair protein RecO family DNA replication, recombination, and repair 2.08 SA1492 nth endonuclease III DNA replication, recombination, and repair 2.03 SA1711 NA DNA-3-methyladenine glycosylase DNA replication, recombination, and repair 1.97 SA1731 dnaI primosomal protein DnaI DNA replication, recombination, and repair 1.87 SA1303 NA competence/damage-inducible protein CinA DNA replication, recombination, and repair 1.72 SA2729 NA integrase/recombinase, core domain family DNA replication, recombination, and repair 1.70 SA0004 NA recF protein DNA replication, recombination, and repair 1.69 SA0880 NA Toprim domain protein Other 1.97
Protein synthesis
SA1369 rpmG1 ribosomal protein L33 Ribosomal proteins: synthesis and modification 12.19 SA1516 rpsA ribosomal protein S1 Ribosomal proteins: synthesis and modification 4.08 SA0815 NA ribosomal subunit interface protein Translation factors 11.79 SA0818 prfB peptide chain release factor 2, authentic frameshift Translation factors 1.67 SA1729 thrS threonyl-tRNA synthetase tRNA aminoacylation 2.42 SA1012 NA ribosomal large subunit pseudouridine synthases, RluD tRNA and rRNA base modification 3.83 subfamily
Protein fate
SA1433 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 11.69 SA2563 NA ATP-dependent Clp protease, putative Degradation of proteins, peptides, and glycopeptides 7.15 SA0979 clpB ATP-dependent Clp protease, ATP-binding subunit Degradation of proteins, peptides, and glycopeptides 5.82 ClpB 8
SA2125 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 4.70 SA0570 clpC ATP-dependent Clp protease, ATP-binding subunit Degradation of proteins, peptides, and glycopeptides 4.48 ClpC, authentic frameshift SA1555 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 4.03 SA2463 pepA2 glutamyl-aminopeptidase Degradation of proteins, peptides, and glycopeptides 3.79 SA1795 pepA1 glutamyl aminopeptidase Degradation of proteins, peptides, and glycopeptides 3.35 SA2322 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 2.91 SA2007 NA peptidase, M20/M25/M40 family, authentic frameshift Degradation of proteins, peptides, and glycopeptides 2.60 SA1588 NA proline dipeptidase Degradation of proteins, peptides, and glycopeptides 2.53 SA0085 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 2.20 SA1937 pepS aminopeptidase PepS Degradation of proteins, peptides, and glycopeptides 2.07 SA0595 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 1.88 SA0833 clpP ATP-dependent Clp protease, proteolytic subunit ClpP Degradation of proteins, peptides, and glycopeptides 1.74 SA1801 NA peptidase, M20/M25/M40 family Degradation of proteins, peptides, and glycopeptides 1.70 SA1721 clpX ATP-dependent Clp protease, ATP-binding subunit Degradation of proteins, peptides, and glycopeptides 1.60 ClpX SA0816 secA preprotein translocase, SecA subunit Protein and peptide secretion and trafficking 1.58 SA1637 dnaK dnaK protein Protein folding and stabilization 3.75 SA2016 groEL chaperonin, 60 kDa Protein folding and stabilization 3.22 SA1271 hslU heat shock protein HslVU, ATPase subunit HslU Protein folding and stabilization 2.55 SA1638 grpE heat shock protein GrpE Protein folding and stabilization 2.31 SA2017 groES chaperonin, 10 kDa Protein folding and stabilization 2.12 SA2385 NA heat shock protein, Hsp20 family Protein folding and stabilization 1.83 SA0957 NA peptidyl-prolyl cis-trans isomerase, cyclophilin-type Protein folding and stabilization 1.74 SA1397 msrA peptide methionine sulfoxide reductase Protein modification and repair 4.42 SA1034 NA lipoate-protein ligase A family protein Protein modification and repair 2.68 SA1591 NA lipoate-protein ligase A family protein Protein modification and repair 1.57
Regulatory functions
SA0766 saeR DNA-binding response regulator SaeR DNA interactions 8.35 SA2193 NA transcriptional regulator, MerR family DNA interactions 7.41 SA0731 NA transcriptional regulator, LysR family DNA interactions 6.08 SA0567 ctsR transcriptional regulator CtsR DNA interactions 5.23 SA0837 gapR gap transcriptional regulator DNA interactions 5.00 SA0404 NA transcriptional regulator, MarR family DNA interactions 4.92 SA1919 NA transcriptional regulator, Fur family DNA interactions 3.24 9
SA1301 NA transcriptional regulator, putative DNA interactions 2.77 SA2389 NA transcriptional regulatory protein DegU, putative DNA interactions 2.65 SA1639 hrcA heat-inducible transcription repressor HrcA DNA interactions 2.51 SA2358 NA DNA-binding response regulator DNA interactions 1.92 SA1541 NA transcriptional regulator, Fur family DNA interactions 1.88 SA2189 NA transcriptional regulator, Sir2 family DNA interactions 1.88 SA1003 NA negative regulator of competence MecA, putative Other 5.90 SA2585 NA regulatory protein, putative Other 3.67 SA2650 NA transcriptional regulator, putative Other 2.40 SA2302 NA transcriptional regulator, putative Other 2.18 SA0307 pfoR perfringolysin O regulator protein Other 2.10 SA0766 saeR DNA-binding response regulator SaeR Protein interactions 8.35 SA2070 kdpD sensor histidine kinase KdpD Protein interactions 5.61 SA1891 NA RNAIII-activating protein TRAP Protein interactions 5.19 SA0765 saeS sensor histidine kinase SaeS Protein interactions 4.42 SA2389 NA transcriptional regulatory protein DegU, putative Protein interactions 2.65 SA1354 NA sensor histidine kinase, putative Protein interactions 2.33 SA0245 lytS sensor histidine kinase LytS Protein interactions 2.21 SA2358 NA DNA-binding response regulator Protein interactions 1.92 SA2390 NA sensory box histidine kinase, putative Protein interactions 1.66 SA1393 NA transcriptional antiterminator LicT, putative RNA interactions 3.53
Signal transduction
SA1457 NA PTS system, IIA component PTS 3.37 SA0224 NA PTS system, IIBC components PTS 2.41 SA2181 lacE PTS system, lactose-specific IIBC components PTS 1.64 SA0766 saeR DNA-binding response regulator SaeR Two-component systems 8.35 SA2070 kdpD sensor histidine kinase KdpD Two-component systems 5.61 SA0765 saeS sensor histidine kinase SaeS Two-component systems 4.42 SA2389 NA transcriptional regulatory protein DegU, putative Two-component systems 2.65 SA1354 NA sensor histidine kinase, putative Two-component systems 2.33 SA0245 lytS sensor histidine kinase LytS Two-component systems 2.21 SA2358 NA DNA-binding response regulator Two-component systems 1.92 SA2390 NA sensory box histidine kinase, putative Two-component systems 1.66
Cell envelope 10
SA1522 NA elastin binding protein, putative Biosynthesis and degradation of surface polysaccharides 5.24 and lipopolysaccharides SA1498 NA glycosyl transferase, group 1 family protein Biosynthesis and degradation of surface polysaccharides 3.45 and lipopolysaccharides SA0611 NA glycosyl transferase, group 1 family protein Biosynthesis and degradation of surface polysaccharides 2.67 and lipopolysaccharides SA0113 NA NAD-dependent epimerase/dehydratase family protein Biosynthesis and degradation of surface polysaccharides 2.04 and lipopolysaccharides SA0138 cap5C capsular polysaccharide biosynthesis protein Cap5C Biosynthesis and degradation of surface polysaccharides 2.01 and lipopolysaccharides SA1804 NA polysaccharide biosynthesis protein Biosynthesis and degradation of surface polysaccharides 1.96 and lipopolysaccharides SA0136 cap5A capsular polysaccharide biosynthesis protein Cap5A Biosynthesis and degradation of surface polysaccharides 1.90 and lipopolysaccharides SA0543 glmU UDP-N-acetylglucosamine pyrophosphorylase Biosynthesis and degradation of surface polysaccharides 1.74 and lipopolysaccharides SA0139 cap5D capsular polysaccharide biosynthesis protein Cap5D Biosynthesis and degradation of surface polysaccharides 1.65 and lipopolysaccharides SA0137 cap5B capsular polysaccharide biosynthesis protein Cap5B Biosynthesis and degradation of surface polysaccharides 1.58 and lipopolysaccharides SA0247 lrgA holin-like protein LrgA Biosynthesis of murein sacculus and peptidoglycan 14.85 SA0248 lrgB lrgB protein Biosynthesis of murein sacculus and peptidoglycan 6.62 SA0238 NA teichoic acid biosynthesis protein, putative Biosynthesis of murein sacculus and peptidoglycan 2.82 SA1161 murI glutamate racemase Biosynthesis of murein sacculus and peptidoglycan 2.69 SA2074 NA D-alanine--D-alanine ligase Biosynthesis of murein sacculus and peptidoglycan 2.39 SA0245 lytS sensor histidine kinase LytS Biosynthesis of murein sacculus and peptidoglycan 2.21 SA2060 alr alanine racemase Biosynthesis of murein sacculus and peptidoglycan 2.17 SA2073 murF UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6- Biosynthesis of murein sacculus and peptidoglycan 1.85 diaminopimelate--D-alanyl-D-alanyl ligase SA1168 efb fibrinogen-binding protein Other 12.45 SA2451 NA amino acid ABC transporter, amino acid-binding protein Other 6.75 SA0995 NA oligopeptide ABC transporter, oligopeptide-binding Other 5.11 protein SA0466 NA membrane protein, putative Other 4.28 SA1762 NA thiol peroxidase, putative Other 2.69 SA0119 sasD cell wall surface anchor family protein Other 2.59 11
SA2272 modA molybdenum ABC transporter, molybdenum-binding Other 2.39 protein ModA SA2668 sasF LPXTG cell wall surface anchor family protein Other 2.35 SA2167 NA iron compound ABC transporter, iron compound- Other 2.29 binding protein SA2539 srtA sortase Other 2.24 SA2505 sasG cell wall surface anchor family protein Other 2.08 SA1424 NA phosphate ABC transporter, phosphate-binding protein Other 2.01
Cellular processes
SA1759 NA universal stress protein family Adaptations to atypical conditions 11.61 SA1010 relA1 GTP pyrophosphokinase Adaptations to atypical conditions 5.43 SA2131 NA Dps family protein Adaptations to atypical conditions 4.64 SA2173 NA alkaline shock protein 23 Adaptations to atypical conditions 2.02 SA1753 NA universal stress protein family Adaptations to atypical conditions 2.00 SA0552 NA general stress protein 13 Adaptations to atypical conditions 1.97 SA0958 NA general stress protein 13 Adaptations to atypical conditions 1.87 SA0551 NA cell-division protein divIC, putative Cell division 3.40 SA2735 NA chromosome partioning protein, ParB family Cell division 1.85 SA0432 NA spoOJ protein Cell division 1.80 SA1368 kataA catalase Detoxification 9.96 SA1610 sodA2 superoxide dismutase Detoxification 4.45 SA0452 ahpC alkyl hydroperoxide reductase, C subunit Detoxification 4.42 SA0451 ahpF alkyl hydroperoxide reductase, subunit F Detoxification 3.92 SA2641 gpxA2 glutathione peroxidase Detoxification 3.45 SA1325 gpxA1 glutathione peroxidase Detoxification 3.17 SA0118 sodA1 superoxide dismutase Detoxification 1.75 SA1645 NA comE operon protein 2 DNA transformation 6.80 SA1003 NA negative regulator of competence MecA, putative DNA transformation 5.90 SA1440 NA xpaC protein, putative Other 4.82 SA2421 hlgC gamma hemolysin, component C Pathogenesis 8.97 SA0766 saeR DNA-binding response regulator SaeR Pathogenesis 8.35 SA2419 hlgA gamma-hemolysin, component A Pathogenesis 6.63 SA1891 NA RNAIII-activating protein TRAP Pathogenesis 5.19 SA0765 saeS sensor histidine kinase SaeS Pathogenesis 4.42 SA2418 NA IgG-binding protein SBI Pathogenesis 3.39 12
SA2389 NA transcriptional regulatory protein DegU, putative Pathogenesis 2.65 SA2006 NA Aerolysin/Leukocidin family protein Pathogenesis 2.36 SA0245 lytS sensor histidine kinase LytS Pathogenesis 2.21 SA0095 spa immunoglobulin G binding protein A precursor Pathogenesis 1.93 SA0105 NA siderophore biosynthesis protein, IucC family Pathogenesis 1.53 SA2422 hlgB gamma hemolysin, component B Toxin production and resistance 13.77 SA2421 hlgC gamma hemolysin, component C Toxin production and resistance 8.97 SA2419 hlgA gamma-hemolysin, component A Toxin production and resistance 6.63 SA1441 NA tellurite resistance protein, putative Toxin production and resistance 5.45 SA0404 NA transcriptional regulator, MarR family Toxin production and resistance 4.92 SA0086 NA drug transporter, putative Toxin production and resistance 3.74 SA2449 NA drug transporter, putative Toxin production and resistance 3.05 SA0163 NA drug transporter, putative Toxin production and resistance 2.48 SA2006 NA Aerolysin/Leukocidin family protein Toxin production and resistance 2.36 SA2570 NA galactoside O-acetyltransferase Toxin production and resistance 2.28 SA2413 NA drug resistance transporter, EmrB/QacA subfamily Toxin production and resistance 1.85
Unknown function
SA0111 NA oxidoreductase, short-chain dehydrogenase/reductase Enzymes of unknown specificity 19.16 family SA2620 NA aminotransferase, class III Enzymes of unknown specificity 12.64 SA2612 NA hydrolase, CocE/NonD family Enzymes of unknown specificity 10.32 SA0569 NA ATP:guanido phosphotransferase family protein Enzymes of unknown specificity 7.07 SA2575 NA aminotransferase, class I Enzymes of unknown specificity 6.20 SA2400 NA acetyltransferase, GNAT family Enzymes of unknown specificity 5.47 SA0959 NA NADH-dependent flavin oxidoreductase, Oye family Enzymes of unknown specificity 5.32 SA2549 NA esterase, putative Enzymes of unknown specificity 5.27 SA1835 NA oxidoreductase, aldo/keto reductase family Enzymes of unknown specificity 5.16 SA2369 NA pyridine nucleotide-disulfide oxidoreductase Enzymes of unknown specificity 4.62 SA1797 NA metallo-beta-lactamase family protein Enzymes of unknown specificity 4.00 SA1543 NA oxidoreductase, aldo/keto reductase family Enzymes of unknown specificity 3.92 SA2192 NA oxidoreductase, aldo/keto reductase family Enzymes of unknown specificity 3.82 SA2597 NA hydrolase, alpha/beta hydrolase fold family Enzymes of unknown specificity 3.67 SA2368 NA acetyltransferase, GNAT family Enzymes of unknown specificity 3.57 SA1116 NA inositol monophosphatase family protein Enzymes of unknown specificity 3.55 SA2667 NA isochorismatase family protein Enzymes of unknown specificity 3.52 13
SA2366 NA acetyltransferase, GNAT family Enzymes of unknown specificity 3.38 SA2321 NA oxidoreductase, short chain dehydrogenase/reductase Enzymes of unknown specificity 3.20 family SA1058 NA aminotransferase, class I Enzymes of unknown specificity 3.04 SA2303 NA inositol monophosphatase family protein Enzymes of unknown specificity 2.88 SA1189 NA acetyltransferase, GNAT family Enzymes of unknown specificity 2.33 SA1981 NA isochorismatase family protein Enzymes of unknown specificity 2.29 SA1365 NA hydrolase, haloacid dehalogenase-like family Enzymes of unknown specificity 2.22 SA0399 NA oxidoreductase, putative Enzymes of unknown specificity 2.20 SA1192 NA conserved hypothetical protein TIGR00006 Enzymes of unknown specificity 2.14 SA1520 NA pyridine nucleotide-disulfide oxidoreductase Enzymes of unknown specificity 2.10 SA0671 NA hydrolase, alpha/beta hydrolase fold family Enzymes of unknown specificity 2.06 SA2594 NA oxidoreductase, short chain dehydrogenase/reductase Enzymes of unknown specificity 1.94 family SA2446 NA epimerase/dehydratase, putative Enzymes of unknown specificity 1.93 SA2021 NA hydrolase, carbon-nitrogen family Enzymes of unknown specificity 1.78 SA0197 NA oxidoreductase, Gfo/Idh/MocA family Enzymes of unknown specificity 1.78 SA0602 NA hydrolase, haloacid dehalogenase-like family Enzymes of unknown specificity 1.68 SA1322 NA hydrolase, alpha/beta hydrolase fold family Enzymes of unknown specificity 1.57 SA0874 NA nitroreductase family protein Enzymes of unknown specificity 1.55 SA0549 NA tetrapyrrole methylase family protein Enzymes of unknown specificity 1.53 SA2278 NA acyl-CoA dehydrogenase-related protein General 7.71 SA1678 NA bacterial luciferase family protein General 7.36 SA1164 NA fibrinogen binding-related protein General 6.48 SA0220 NA flavohemoprotein, putative General 6.38 SA1187 NA antibacterial protein (phenol soluble modulin) General 5.18 SA1169 NA fibrinogen-binding protein precursor-related protein General 4.83 SA2660 isaB immunodominant antigen B General 4.28 SA2522 NA DedA family protein General 4.25 SA1768 NA GAF domain protein General 4.25 SA0408 NA glyoxalase family protein General 4.21 SA2533 NA glyoxalase family protein General 3.41 SA0872 NA OsmC/Ohr family protein General 3.05 SA0410 NA FMN reductase-related protein General 3.03 SA1186 NA antibacterial protein (phenol soluble modulin) General 2.92 SA1162 NA HAM1 protein General 2.79 SA1712 NA abrB protein General 2.53 14
abrB protein, putative SA2156 NA ATP-binding protein, Mrp/Nbp35 family General 2.53 SA0821 NA HD domain protein General 2.43 SA1733 NA ATP cone domain protein General 2.41 SA0014 NA DHH subfamily 1 protein General 2.28 SA0550 NA S4 domain protein General 2.27 SA1628 NA PhoH family protein General 2.17 SA0658 NA HD domain protein General 2.14 SA1771 NA OsmC/Ohr family protein General 2.12 SA1621 NA CBS domain protein General 2.09 SA1220 NA fibronectin/fibrinogen binding-related protein General 2.06 SA1207 NA glyoxalase family protein General 2.04 SA1326 NA GTP-binding protein, putative General 2.00 SA2195 NA M23/M37 peptidase domain protein General 1.95 SA2333 NA YnfA family protein General 1.91 SA1300 NA ACT domain protein General 1.90 SA0618 NA SIS domain protein General 1.90 SA0708 NA DAK2 domain protein General 1.90 SA1592 NA rhodanese-like domain protein General 1.88 SA1894 NA HIT family protein General 1.85 SA0239 NA TagF domain protein General 1.85 SA0707 NA dihydroxyacetone kinase family protein General 1.84 SA0541 spoVG spoVG protein General 1.78 SA0921 NA CBS domain protein General 1.64
Hypothetical proteins
SA0768 NA conserved hypothetical protein Conserved 19.26 SA2550 NA conserved hypothetical protein Conserved 16.05 SA0467 NA conserved hypothetical protein Conserved 12.12 SA1026 NA conserved hypothetical protein Conserved 10.73 SA2551 NA conserved hypothetical protein TIGR00051 Conserved 8.35 SA0767 NA conserved hypothetical protein Conserved 8.24 SA1006 NA conserved hypothetical protein Conserved 7.65 SA0597 NA conserved hypothetical protein Conserved 7.39 SA1438 NA conserved hypothetical protein Conserved 5.89 SA2241 NA conserved hypothetical protein Conserved 5.61 15
SA0161 NA conserved hypothetical protein Conserved 5.53 SA1992 NA conserved hypothetical protein Conserved 5.51 SA1226 NA conserved hypothetical protein Conserved 5.48 SA2124 NA conserved hypothetical protein Conserved 4.96 SA0568 NA conserved hypothetical protein Conserved 4.90 SA0730 NA conserved hypothetical protein Conserved 4.69 SA1358 NA conserved hypothetical protein Conserved 4.68 SA0409 NA conserved hypothetical protein Conserved 4.55 SA2179 NA conserved hypothetical protein Conserved 4.47 SA1011 NA conserved hypothetical protein Conserved 4.20 SA1670 NA conserved hypothetical protein Conserved 4.11 SA1927 NA conserved hypothetical protein Conserved 3.93 SA2710 NA conserved hypothetical protein Conserved 3.92 SA1190 NA conserved hypothetical protein Conserved 3.76 SA1113 NA conserved hypothetical protein Conserved 3.76 SA1009 NA conserved hypothetical protein Conserved 3.69 SA2132 NA conserved hypothetical protein Conserved 3.67 SA1485 NA conserved hypothetical protein Conserved 3.65 SA0427 NA conserved hypothetical protein Conserved 3.62 SA0444 NA conserved hypothetical protein Conserved 3.60 SA2456 NA conserved hypothetical protein Conserved 3.59 SA2143 NA conserved hypothetical protein Conserved 3.56 SA0669 NA conserved hypothetical protein Conserved 3.55 SA1486 NA conserved hypothetical protein Conserved 3.39 SA0943 NA conserved hypothetical protein Conserved 3.35 SA1163 NA conserved hypothetical protein TIGR00040 Conserved 3.29 SA2439 NA conserved hypothetical protein Conserved 3.23 SA0456 NA conserved hypothetical protein Conserved 3.23 SA1672 NA conserved hypothetical protein Conserved 3.16 SA0495 NA conserved hypothetical protein Conserved 3.09 SA0922 NA conserved hypothetical protein Conserved 3.04 SA0614 NA conserved hypothetical protein Conserved 3.01 SA1754 NA conserved hypothetical protein, authentic frameshift Conserved 2.92 SA1671 NA conserved hypothetical protein TIGR00250 Conserved 2.83 SA0925 NA conserved hypothetical protein Conserved 2.82 SA0129 NA conserved hypothetical protein Conserved 2.79 SA1895 NA conserved hypothetical protein Conserved 2.74 16
SA1557 NA conserved hypothetical protein Conserved 2.69 SA2365 NA conserved hypothetical protein Conserved 2.69 SA0924 NA conserved hypothetical protein Conserved 2.66 SA2063 NA conserved hypothetical protein Conserved 2.60 SA1101 NA conserved hypothetical protein Conserved 2.56 SA0811 NA conserved hypothetical protein TIGR00257 Conserved 2.49 SA0942 NA conserved hypothetical protein Conserved 2.48 SA2391 NA conserved hypothetical protein Conserved 2.47 SA1975 NA conserved hypothetical protein Conserved 2.44 SA1128 NA conserved hypothetical protein Conserved 2.40 SA1688 NA conserved hypothetical protein TIGR00256 Conserved 2.40 SA0709 NA conserved hypothetical protein Conserved 2.39 SA0445 NA conserved hypothetical protein Conserved 2.39 SA0615 NA conserved hypothetical protein Conserved 2.37 SA0314 NA conserved hypothetical protein Conserved 2.35 SA2330 NA conserved hypothetical protein Conserved 2.25 SA2174 NA conserved hypothetical protein Conserved 2.21 SA2711 NA conserved hypothetical protein Conserved 2.21 SA0457 NA conserved hypothetical protein Conserved 2.21 SA1620 NA conserved hypothetical protein Conserved 2.20 SA0776 NA conserved hypothetical protein TIGR00370 Conserved 2.17 SA2343 NA conserved hypothetical protein Conserved 2.15 SA2175 NA conserved hypothetical protein Conserved 2.11 SA1500 NA conserved hypothetical protein Conserved 2.10 SA2364 NA conserved hypothetical protein Conserved 2.09 SA1480 NA conserved hypothetical protein Conserved 2.08 SA0787 NA conserved hypothetical protein TIGR00147 Conserved 2.03 SA1418 NA conserved hypothetical protein Conserved 2.02 SA1402 NA glutamyl aminopeptidase, putative Conserved 2.01 SA0727 NA conserved hypothetical protein TIGR01033 Conserved 1.99 SA1387 NA conserved hypothetical protein Conserved 1.95 SA2020 NA conserved hypothetical protein Conserved 1.93 SA1499 NA conserved hypothetical protein Conserved 1.91 SA0446 NA conserved hypothetical protein Conserved 1.89 SA0831 NA conserved hypothetical protein Conserved 1.89 SA1558 NA conserved hypothetical protein Conserved 1.87 SA0577 NA conserved hypothetical protein Conserved 1.87 17
SA1089 NA conserved hypothetical protein Conserved 1.87 SA0013 NA conserved hypothetical protein Conserved 1.86 SA1395 NA conserved hypothetical protein Conserved 1.85 SA0065 NA conserved hypothetical protein Conserved 1.82 SA0218 NA conserved hypothetical protein Conserved 1.82 SA1129 NA conserved hypothetical protein Conserved 1.81 SA1503 NA conserved hypothetical protein Conserved 1.81 SA2609 NA conserved hypothetical protein Conserved 1.80 SA0647 NA conserved hypothetical protein Conserved 1.78 SA2162 NA conserved hypothetical protein Conserved 1.73 SA1200 NA conserved hypothetical protein TIGR00726 Conserved 1.71 SA1923 NA conserved hypothetical protein Conserved 1.69 SA1327 NA conserved hypothetical protein Conserved 1.67 SA2106 NA conserved hypothetical protein TIGR01440 Conserved 1.66 SA2288 NA conserved hypothetical protein Conserved 1.65 SA1090 NA conserved hypothetical protein Conserved 1.63 SA1456 NA conserved hypothetical protein Conserved 1.60 SA2723 NA conserved hypothetical protein Conserved 1.57 SA2300 NA conserved hypothetical protein Conserved 1.55 SA0692 NA conserved hypothetical protein Conserved 1.53 SA2304 NA conserved domain protein Domain 2.44 SA0396 NA conserved domain protein Domain 1.87
Downregulated Genes
Amino acid biosynthesis
SA1652 aroE shikimate 5-dehydrogenase Aromatic amino acid family -2.01 SA0503 NA trans-sulfuration enzyme family protein Aspartate family -2.23 SA2702 hisD histidinol dehydrogenase Histidine family -5.21 SA0100 NA cysteine synthase/cystathionine beta-synthase family Serine family -4.01 protein SA0503 NA trans-sulfuration enzyme family protein Serine family -2.23
Purines, pyrimidines, nucleosides, and nucleotides
SA0792 NA ribonucleoside-diphosphate reductase 2, alpha subunit 2'-Deoxyribonucleotide metabolism -2.01 18
SA2635 nrdD anaerobic ribonucleoside-triphosphate reductase 2'-Deoxyribonucleotide metabolism -1.81 SA1371 guaC GMP reductase Nucleotide and nucleoside interconversions -5.14 SA2111 tdk thymidine kinase Nucleotide and nucleoside interconversions -4.90 SA0604 NA deoxynucleoside kinase family protein Nucleotide and nucleoside interconversions -4.23 SA1509 NA nucleoside diphosphate kinase Nucleotide and nucleoside interconversions -3.74 SA2218 adk adenylate kinase Nucleotide and nucleoside interconversions -2.39 SA0603 NA deoxynucleoside kinase family protein Nucleotide and nucleoside interconversions -2.24 SA1277 pyrH uridylate kinase Nucleotide and nucleoside interconversions -1.74 SA1221 gmk guanylate kinase Nucleotide and nucleoside interconversions -1.72 SA1518 cmk cytidylate kinase Nucleotide and nucleoside interconversions -1.69 SA0024 sasH 5-nucleotidase family protein Other -2.91 SA1074 purK phosphoribosylaminoimidazole carboxylase, ATPase Purine ribonucleotide biosynthesis -15.24 subunit SA1969 purB adenylosuccinate lyase Purine ribonucleotide biosynthesis -9.78 SA1079 purF amidophosphoribosyltransferase Purine ribonucleotide biosynthesis -7.76 SA1077 purQ phosphoribosylformylglycinamidine synthase I Purine ribonucleotide biosynthesis -7.75 SA1073 purE phosphoribosylaminoimidazole carboxylase, catalytic Purine ribonucleotide biosynthesis -6.75 subunit SA1075 purC phosphoribosylaminoimidazole-succinocarboxamide Purine ribonucleotide biosynthesis -6.64 synthase SA1080 purM phosphoribosylformylglycinamidine cyclo-ligase Purine ribonucleotide biosynthesis -5.23 SA1078 purL phosphoribosylformylglycinamidine synthase II Purine ribonucleotide biosynthesis -4.75 SA0460 guaB inosine-5-monophosphate dehydrogenase Purine ribonucleotide biosynthesis -3.45 SA0018 purA adenylosuccinate synthetase Purine ribonucleotide biosynthesis -3.41 SA0461 guaA GMP synthase Purine ribonucleotide biosynthesis -3.39 SA1081 purN phosphoribosylglycinamide formyltransferase Purine ribonucleotide biosynthesis -2.72 SA1083 purD phosphoribosylamine--glycine ligase Purine ribonucleotide biosynthesis -1.90 SA0544 prsA ribose-phosphate pyrophosphokinase Purine ribonucleotide biosynthesis -1.77 SA1076 purS phosphoribosylformylglycinamidine synthase, PurS Purine ribonucleotide biosynthesis -1.75 protein SA1082 purH phosphoribosylaminoimidazolecarboxamide Purine ribonucleotide biosynthesis -1.66 formyltransferase/IMP cyclohydrolase SA2119 pyrG CTP synthase Pyrimidine ribonucleotide biosynthesis -4.91 SA2606 pyrD dihydroorotate dehydrogenase Pyrimidine ribonucleotide biosynthesis -4.62 SA1215 carB carbamoyl-phosphate synthase, large subunit Pyrimidine ribonucleotide biosynthesis -4.12 SA1212 pyrB aspartate carbamoyltransferase Pyrimidine ribonucleotide biosynthesis -2.73 SA1216 pyrF orotidine 5-phosphate decarboxylase Pyrimidine ribonucleotide biosynthesis -2.14 19
SA1214 carA carbamoyl-phosphate synthase, small subunit Pyrimidine ribonucleotide biosynthesis -1.66 SA0458 xpt xanthine phosphoribosyltransferase Salvage of nucleosides and nucleotides -6.39 SA0225 NA inosine-uridine preferring nucleoside hydrolase Salvage of nucleosides and nucleotides -2.26 SA1690 apt adenine phosphoribosyltransferase Salvage of nucleosides and nucleotides -1.78 SA0554 hpt hypoxanthine phosphoribosyltransferase Salvage of nucleosides and nucleotides -1.71
Fatty acid and phospholipid metabolism
SA1280 cdsA phosphatidate cytidylyltransferase Biosynthesis -7.42 SA1299 NA acetoacetyl-CoA reductase, putative Biosynthesis -4.82 SA0987 fabH 3-oxoacyl-(acyl-carrier-protein) synthase III Biosynthesis -2.96 SA1244 fabD malonyl CoA-acyl carrier protein transacylase Biosynthesis -2.67 SA1748 accD acetyl-CoA carboxylase, carboxyl transferase, beta Biosynthesis -2.27 subunit SA1572 accB acetyl-CoA carboxylase, biotin carboxyl carrier protein Biosynthesis -2.20 SA0426 NA acetyl-CoA acetyltransferase Biosynthesis -2.12 SA1571 accC acetyl-CoA carboxylase, biotin carboxylase Biosynthesis -1.92
Biosynthesis of cofactors, prosthetic groups, and carriers
SA2427 bioA adenosylmethionine--8-amino-7-oxononanoate Biotin -1.93 aminotransferase SA1709 folC folylpolyglutamate synthase/dihydrofolate synthase Folic acid -2.62 SA1124 ctaA cytochrome oxidase assembly protein Heme, porphyrin, and cobalamin -10.78 SA1125 ctaB protoheme IX farnesyltransferase Heme, porphyrin, and cobalamin -5.39 SA1717 hemC porphobilinogen deaminase Heme, porphyrin, and cobalamin -2.43 SA2598 NA cobalamin synthesis protein, putative Heme, porphyrin, and cobalamin -2.03 SA1049 menA 1,4-dihydroxy-2-naphthoate octaprenyltransferase Menaquinone and ubiquinone -3.04 SA1052 menD 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic Menaquinone and ubiquinone -2.88 acid synthase/2-oxoglutarate decarboxylase SA1566 ispA geranyltranstransferase Menaquinone and ubiquinone -2.49 SA0637 mvaD mevalonate diphosphate decarboxylase Other -3.79 SA2561 NA hydroxymethylglutaryl-CoA synthase Other -3.48 SA0636 mvk mevalonate kinase Other -2.37 SA1279 uppS undecaprenyl diphosphate synthase Other -2.17 SA2579 NA phytoene dehydrogenase Other -2.17 SA0638 NA phosphomevalonate kinase Other -2.14 20
SA0172 entB isochorismatase Other -1.70 SA2341 fni isopentenyl diphosphate isomerase Other -1.54 SA0564 NA pyridoxine biosynthesis protein Pyridoxine -7.82 SA1817 ribH riboflavin synthase, beta subunit Riboflavin, FMN, and FAD -1.70 SA1818 ribBA 3,4-dihydroxy-2-butanone-4-phosphate synthase/GTP Riboflavin, FMN, and FAD -1.57 cyclohydrolase II SA2085 thiD2 phosphomethylpyrimidine kinase Thiamine -5.33 SA1764 thiI thiamine biosynthesis protein ThiI Thiamine -4.00 SA2084 thiM hydroxyethylthiazole kinase Thiamine -2.33
Central intermediary metabolism
SA2145 glmS glucosamine--fructose-6-phosphate aminotransferase Amino sugars -5.49 (isomerizing) SA1837 metK S-adenosylmethionine synthetase One-carbon metabolism -5.43 SA2535 NA D-isomer specific 2-hydroxyacid dehydrogenase family Other -5.12 protein SA0637 mvaD mevalonate diphosphate decarboxylase Other -3.79 SA2561 NA hydroxymethylglutaryl-CoA synthase Other -3.48 SA2576 crtN dehydrosqualene desaturase Other -2.43 SA0636 mvk mevalonate kinase Other -2.37 SA0638 NA phosphomevalonate kinase Other -2.14 SA1760 ackA acetate kinase Other -2.10 SA1655 mtn 5-methylthioadenosine/S-adenosylhomocysteine Other -1.65 nucleosidase SA0173 ipdC indole-3-pyruvate decarboxylase Other -1.57
Energy metabolism
SA2544 sdaAA L-serine dehydratase, iron-sulfur-dependent, alpha Amino acids and amines -3.15 subunit SA2545 sdaAB L-serine dehydratase, iron-sulfur-dependent, beta Amino acids and amines -3.08 subunit SA2569 NA delta-1-pyrroline-5-carboxylate dehydrogenase, putative Amino acids and amines -2.36 SA1560 NA 2-oxoisovalerate dehydrogenase, E2 component, Amino acids and amines -2.30 dihydrolipoamide acetyltransferase SA1758 ald2 alanine dehydrogenase Amino acids and amines -2.04 21
SA1594 NA glycine cleavage system P protein, subunit 1 Amino acids and amines -1.79 SA0008 hutH histidine ammonia-lyase Amino acids and amines -1.67 SA1593 NA glycine cleavage system P protein, subunit 2 Amino acids and amines -1.67 SA2099 atpF ATP synthase F0, B subunit ATP-proton motive force interconversion -3.48 SA2101 atpB ATP synthase F0, A subunit ATP-proton motive force interconversion -3.20 SA2098 atpH ATP synthase F1, delta subunit ATP-proton motive force interconversion -3.00 SA2097 atpA ATP synthase F1, alpha subunit ATP-proton motive force interconversion -2.76 SA2095 atpD ATP synthase F1, beta subunit ATP-proton motive force interconversion -2.46 SA2096 atpG ATP synthase F1, gamma subunit ATP-proton motive force interconversion -2.41 SA2100 atpE ATP synthase F0, C subunit ATP-proton motive force interconversion -1.62 SA1551 malA alpha-glucosidase Biosynthesis and degradation of polysaccharides -2.40 SA1158 sdhC succinate dehydrogenase, cytochrome b558 subunit Electron transport -2.98 SA1095 cydB cytochrome d ubiquinol oxidase, subunit II Electron transport -2.16 SA1094 cydA cytochrome d ubiquinol oxidase, subunit I Electron transport -1.89 SA1914 NA iron-sulfur cluster-binding protein, putative Electron transport -1.73 SA1760 ackA acetate kinase Fermentation -2.10 SA0135 NA alcohol dehydrogenase, iron-containing Fermentation -1.76 SA1838 pckA phosphoenolpyruvate carboxykinase (ATP) Glycolysis/gluconeogenesis -11.48 SA1734 gapA2 glyceraldehyde 3-phosphate dehydrogenase Glycolysis/gluconeogenesis -2.23 SA1604 glk glucokinase Glycolysis/gluconeogenesis -1.87 SA2527 NA fructose-1,6-bisphosphatase, putative Glycolysis/gluconeogenesis -1.53 SA1320 glpK glycerol kinase Other -5.83 SA1749 NA NADP-dependent malic enzyme, putative Other -2.86 SA1514 gpsA glycerol-3-phosphate dehydrogenase, NAD-dependent Other -1.92 SA1103 pdhB pyruvate dehydrogenase complex E1 component, beta Pyruvate dehydrogenase -1.73 subunit SA2135 manA1 mannose-6-phosphate isomerase, class I Sugars -6.41 SA0253 rbsK ribokinase Sugars -2.09 SA1158 sdhC succinate dehydrogenase, cytochrome b558 subunit TCA cycle -2.98 SA1449 sucA 2-oxoglutarate dehydrogenase, E1 component TCA cycle -2.89 SA1749 NA NADP-dependent malic enzyme, putative TCA cycle -2.86 SA1160 sdhB succinate dehydrogenase, iron-sulfur protein TCA cycle -2.75 SA1385 acnA aconitate hydratase TCA cycle -2.39 SA1263 sucD succinyl-CoA synthase, alpha subunit TCA cycle -1.52 SA1159 sdhA succinate dehydrogenase, flavoprotein subunit TCA cycle -1.51
Transport and binding proteins 22
SA2382 NA proton/sodium-glutamate symport protein Amino acids, peptides and amines -7.71 SA1367 NA amino acid permease Amino acids, peptides and amines -5.27 SA2632 cudT osmoprotectant transporter, BCCT family Amino acids, peptides and amines -5.02 SA1108 NA spermidine/putrescine ABC transporter, ATP-binding Amino acids, peptides and amines -4.75 protein SA2441 NA amino acid permease Amino acids, peptides and amines -4.61 SA1111 NA spermidine/putrescine ABC transporter, Amino acids, peptides and amines -4.44 spermidine/putrescine-binding protein SA0620 proP osmoprotectant proline transporter Amino acids, peptides and amines -4.13 SA1110 NA spermidine/putrescine ABC transporter, permease Amino acids, peptides and amines -3.22 protein SA0171 brnQ1 branched-chain amino acid transport system II carrier Amino acids, peptides and amines -2.92 protein SA0630 NA amino acid permease Amino acids, peptides and amines -2.85 SA1392 NA sodium:alanine symporter family protein Amino acids, peptides and amines -2.57 SA2472 NA peptide ABC transporter, ATP-binding protein Amino acids, peptides and amines -2.12 SA1743 NA amino acid permease Amino acids, peptides and amines -1.99 SA2176 opuD2 osmoprotectant transporter, BCCT family Amino acids, peptides and amines -1.96 SA2473 NA peptide ABC transporter, ATP-binding protein Amino acids, peptides and amines -1.95 SA2340 gltS sodium:glutamate symporter Amino acids, peptides and amines -1.81 SA2309 NA amino acid permease Amino acids, peptides and amines -1.68 SA1443 brnQ3 branched-chain amino acid transport system II carrier Amino acids, peptides and amines -1.64 protein SA0722 NA phosphate transporter family protein Anions -7.39 SA2401 NA formate/nitrite transporter family protein Anions -3.06 SA0301 NA formate/nitrite transporter family protein Anions -2.40 SA0088 NA Na/Pi cotransporter family protein Anions -1.55 SA2376 NA PTS system, sucrose-specific IIBC components, Carbohydrates, organic alcohols, and acids -10.57 putative SA2316 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids -8.25 SA0093 NA L-lactate permease Carbohydrates, organic alcohols, and acids -6.89 SA0516 NA PTS system, IIBC components Carbohydrates, organic alcohols, and acids -3.25 SA0175 NA PTS system, IIABC components Carbohydrates, organic alcohols, and acids -3.21 SA2246 NA sugar transporter, putative Carbohydrates, organic alcohols, and acids -3.19 SA2401 NA formate/nitrite transporter family protein Carbohydrates, organic alcohols, and acids -3.06 SA0301 NA formate/nitrite transporter family protein Carbohydrates, organic alcohols, and acids -2.40 23
SA2363 NA L-lactate permease Carbohydrates, organic alcohols, and acids -2.27 SA2552 NA PTS system, IIABC components Carbohydrates, organic alcohols, and acids -2.13 SA0192 NA maltose ABC transporter, ATP-binding protein, putative Carbohydrates, organic alcohols, and acids -2.01 SA0193 NA maltose ABC transporter, maltose-binding protein, Carbohydrates, organic alcohols, and acids -1.81 putative SA1917 NA PTS system, IIC component Carbohydrates, organic alcohols, and acids -1.77 SA2514 gntP gluconate transporter, permease protein Carbohydrates, organic alcohols, and acids -1.76 SA0254 NA ribose transport protein Carbohydrates, organic alcohols, and acids -1.60 SA1084 NA cobalt transport family protein Cations and iron carrying compounds -3.92 SA1979 NA sodium-dependent transporter Cations and iron carrying compounds -3.40 SA0949 mnhG Na+/H+ antiporter, MnhG component Cations and iron carrying compounds -3.38 SA2375 NA transporter, CorA family Cations and iron carrying compounds -2.91 SA0951 mnhE Na+/H+ antiporter, MnhE component Cations and iron carrying compounds -2.85 SA0097 sirC iron compound ABC transporter, permease protein SirC Cations and iron carrying compounds -2.66 SA0098 sirB iron compound ABC transporter, permease protein SirB Cations and iron carrying compounds -2.35 SA2442 NA Na+/H+ antiporter, putative Cations and iron carrying compounds -2.19 SA0953 mnhC Na+/H+ antiporter, MnhC component Cations and iron carrying compounds -2.08 SA1096 NA TrkA potassium uptake family protein Cations and iron carrying compounds -2.03 SA0682 NA Na+/H+ antiporter, MnhD component, putative Cations and iron carrying compounds -1.92 SA0952 mnhD Na+/H+ antiporter, MnhD component Cations and iron carrying compounds -1.90 SA0799 NA transferrin receptor Cations and iron carrying compounds -1.89 SA0099 sirA iron compound ABC transporter, iron compound- Cations and iron carrying compounds -1.81 binding protein SirA SA0203 NA iron compound ABC transporter, iron compound- Cations and iron carrying compounds -1.73 binding protein, putative SA0681 NA Na+/H+ antiporter, MnhC component, putative Cations and iron carrying compounds -1.68 SA0955 mnhA Na+/H+ antiporter, MnhA component Cations and iron carrying compounds -1.65 SA0950 mnhF Na+/H+ antiporter, MnhF component Cations and iron carrying compounds -1.62 SA0954 mnhB Na+/H+ antiporter, MnhB component Cations and iron carrying compounds -1.56 SA0088 NA Na/Pi cotransporter family protein Cations and iron carrying compounds -1.55 SA2242 NA xanthine/uracil permease family protein Nucleosides, purines and pyrimidines -15.03 SA0459 pbuX xanthine permease Nucleosides, purines and pyrimidines -6.06 SA0566 nupC nucleoside permease NupC Nucleosides, purines and pyrimidines -4.88 SA0701 NA nucleoside permease NupC, putative Nucleosides, purines and pyrimidines -4.65 SA0407 glpT glycerol-3-phosphate transporter Other -4.08 SA2437 bcr bicyclomycin resistance protein Other -3.75 SA2257 NA drug transporter, putative Other -3.10 24
SA2460 NA drug transporter, putative Other -2.97 SA1823 arsB arsenical pump membrane protein Other -1.93 SA0122 NA tetracycline resistance protein, putative Other -1.91 SA0695 NA tagG protein, teichoic acid ABC transporter protein, Other -1.74 putative SA1809 NA hypothetical protein Other -1.73 SA2347 NA drug resistance transporter, EmrB/QacA subfamily Other -1.72 SA0117 NA polysaccharide extrusion protein Other -1.65 SA1319 glpF glycerol uptake facilitator protein Other -1.61 SA1996 NA ABC transporter, ATP-binding protein Unknown substrate -6.87 SA2521 NA transporter, putative Unknown substrate -6.69 SA0689 NA ABC transporter, permease protein Unknown substrate -5.72 SA0688 NA ABC transporter, substrate-binding protein Unknown substrate -5.09 SA1085 NA ABC transporter, ATP-binding protein Unknown substrate -4.21 SA1994 NA ABC transporter, ATP-binding protein Unknown substrate -3.99 SA0690 NA ABC transporter, ATP-binding protein Unknown substrate -3.63 SA1979 NA sodium-dependent transporter Unknown substrate -3.40 SA2335 NA ABC transporter, ATP-binding protein Unknown substrate -2.78 SA2403 NA ABC transporter, substrate-binding protein Unknown substrate -2.56 SA1893 NA ABC transporter, ATP-binding protein Unknown substrate -2.01 SA0779 NA ABC transporter, ATP-binding protein Unknown substrate -1.86 SA0217 NA ABC transporter, substrate-binding protein Unknown substrate -1.63 SA1613 NA ABC transporter, ATP-binding protein Unknown substrate -1.51
DNA metabolism
SA1150 rnhC ribonuclease HIII DNA replication, recombination, and repair -3.37 SA0001 dnaA chromosomal replication initiator protein DnaA DNA replication, recombination, and repair -2.92 SA1489 recU recombination protein U DNA replication, recombination, and repair -2.16 SA1316 hexB DNA mismatch repair protein HexB DNA replication, recombination, and repair -1.97 SA0343 NA prophage L54a, replicative DNA helicase, putative DNA replication, recombination, and repair -1.92 SA1966 pcrA ATP-dependent DNA helicase PcrA, FRAMESHIFT DNA replication, recombination, and repair -1.90 SA0438 ssb2 single-stranded DNA-binding protein DNA replication, recombination, and repair -1.89 SA0823 uvrB excinuclease ABC, B subunit DNA replication, recombination, and repair -1.83 SA1619 dnaG DNA primase DNA replication, recombination, and repair -1.82 SA1841 NA MutT/nudix family protein DNA replication, recombination, and repair -1.81 SA2339 NA DNA-3-methyladenine glycosylase DNA replication, recombination, and repair -1.74 25
SA0002 dnaN DNA polymerase III, beta subunit DNA replication, recombination, and repair -1.74 SA1965 ligA DNA ligase, NAD-dependent DNA replication, recombination, and repair -1.68 SA1283 NA DNA polymerase III, alpha subunit, Gram-positive type DNA replication, recombination, and repair -1.64 SA1696 ruvB Holliday junction DNA helicase RuvB DNA replication, recombination, and repair -1.50 SA0180 NA type I restriction-modification enzyme, R subunit Restriction/modification -2.58 SA1862 hsdM2 type I restriction-modification system, M subunit Restriction/modification -1.69
Transcription
SA0589 rpoC DNA-directed RNA polymerase, beta subunit DNA-dependent RNA polymerase -4.12 SA0588 rpoB DNA-directed RNA polymerase, beta subunit DNA-dependent RNA polymerase -2.97 SA2120 rpoE DNA-directed RNA polymerase, delta subunit DNA-dependent RNA polymerase -2.12 SA2072 NA ATP-dependent RNA helicase, DEAD/DEAH box Other -5.30 family SA1615 NA ATP-dependent RNA helicase, DEAD/DEAH box Other -2.12 family SA1941 NA ribonuclease BN, putative RNA processing -4.61 SA2739 rnpA ribonuclease P protein component RNA processing -1.99 SA1255 rimM 16S rRNA processing protein RimM RNA processing -1.83 SA2054 rpoF RNA polymerase sigma-37 factor Transcription factors -6.13 SA2113 rho transcription termination factor Rho Transcription factors -3.97 SA2055 rsbW anti-sigma B factor Transcription factors -2.66 SA0582 nusG transcription antitermination protein NusG Transcription factors -2.59 SA1618 rpoD RNA polymerase sigma-70 factor Transcription factors -1.83
Protein synthesis
SA0583 rplK ribosomal protein L11 Ribosomal proteins: synthesis and modification -10.19 SA2239 rplC ribosomal protein L3 Ribosomal proteins: synthesis and modification -9.44 SA2240 rpsJ ribosomal protein S10 Ribosomal proteins: synthesis and modification -7.91 SA0586 rplL ribosomal protein L7/L12 Ribosomal proteins: synthesis and modification -7.84 SA2238 rplD ribosomal protein L4 Ribosomal proteins: synthesis and modification -7.59 SA0592 rpsG ribosomal protein S7 Ribosomal proteins: synthesis and modification -7.36 SA0584 rplA ribosomal protein L1 Ribosomal proteins: synthesis and modification -7.34 SA2206 rpsI ribosomal protein S9 Ribosomal proteins: synthesis and modification -6.70 SA1725 rplT ribosomal protein L20 Ribosomal proteins: synthesis and modification -6.54 SA0590 NA 30S ribosomal protein L7 Ae Ribosomal proteins: synthesis and modification -6.05 26
SA0585 rplJ ribosomal protein L10 Ribosomal proteins: synthesis and modification -5.62 SA2237 rplW ribosomal protein L23 Ribosomal proteins: synthesis and modification -5.26 SA0437 rpsF ribosomal protein S6 Ribosomal proteins: synthesis and modification -5.08 SA0591 rpsL ribosomal protein S12 Ribosomal proteins: synthesis and modification -4.89 SA1702 rplU ribosomal protein L21 Ribosomal proteins: synthesis and modification -4.63 SA2235 rpsS ribosomal protein S19 Ribosomal proteins: synthesis and modification -4.45 SA0439 rpsR ribosomal protein S18 Ribosomal proteins: synthesis and modification -4.40 SA2234 rplV ribosomal protein L22 Ribosomal proteins: synthesis and modification -4.23 SA1257 rplS ribosomal protein L19 Ribosomal proteins: synthesis and modification -4.21 SA2236 rplB ribosomal protein L2 Ribosomal proteins: synthesis and modification -4.05 SA2039 NA ribosomal-protein-alanine acetyltransferase, putative Ribosomal proteins: synthesis and modification -3.94 SA1726 rpmI ribosomal protein L35 Ribosomal proteins: synthesis and modification -3.83 SA1254 rpsP ribosomal protein S16 Ribosomal proteins: synthesis and modification -3.79 SA2233 rpsC ribosomal protein S3 Ribosomal proteins: synthesis and modification -3.71 SA2207 rplM ribosomal protein L13 Ribosomal proteins: synthesis and modification -3.67 SA1642 rpsT ribosomal protein S20 Ribosomal proteins: synthesis and modification -3.44 SA2229 rplN ribosomal protein L14 Ribosomal proteins: synthesis and modification -3.24 SA1769 rpsD ribosomal protein S4 Ribosomal proteins: synthesis and modification -3.20 SA2228 rplX ribosomal protein L24 Ribosomal proteins: synthesis and modification -3.08 SA2227 rplE ribosomal protein L5 Ribosomal proteins: synthesis and modification -2.87 SA0545 rplY ribosomal Protein L25 Ribosomal proteins: synthesis and modification -2.87 SA2231 rpmC ribosomal protein L29 Ribosomal proteins: synthesis and modification -2.69 SA2225 rpsH ribosomal protein S8 Ribosomal proteins: synthesis and modification -2.62 SA2223 rplR ribosomal protein L18 Ribosomal proteins: synthesis and modification -2.59 SA2215 rpsM ribosomal protein S13/S18 Ribosomal proteins: synthesis and modification -2.54 SA2214 rpsK ribosomal protein S11 Ribosomal proteins: synthesis and modification -2.51 SA2740 rpmH ribosomal protein L34 Ribosomal proteins: synthesis and modification -2.50 SA2232 rplP ribosomal protein L16 Ribosomal proteins: synthesis and modification -2.45 SA2224 rplF ribosomal protein L6 Ribosomal proteins: synthesis and modification -2.28 SA2212 rplQ ribosomal protein L17 Ribosomal proteins: synthesis and modification -2.21 SA2230 rpsQ ribosomal protein S17 Ribosomal proteins: synthesis and modification -2.11 SA2220 rplO ribosomal protein L15 Ribosomal proteins: synthesis and modification -2.09 SA1274 rpsB ribosomal protein S2 Ribosomal proteins: synthesis and modification -1.98 SA2226 rpsN2 ribosomal protein S14 Ribosomal proteins: synthesis and modification -1.85 SA1727 infC translation initiation factor IF-3 Translation factors -8.22 SA2110 prfA peptide chain release factor 1 Translation factors -6.77 SA0593 fusA translation elongation factor G Translation factors -2.71 27
SA1278 frr ribosome recycling factor Translation factors -2.07 SA1587 efp translation elongation factor P Translation factors -1.72 SA1288 infB translation initiation factor IF-2 Translation factors -1.65 SA1960 gatB glutamyl-tRNA(Gln) amidotransferase, B subunit tRNA aminoacylation -5.94 SA1961 gatA glutamyl-tRNA(Gln) amidotransferase, A subunit tRNA aminoacylation -5.34 SA1778 tyrS tyrosyl-tRNA synthetase tRNA aminoacylation -4.76 SA1962 gatC glutamyl-tRNA(Gln) amidotransferase, C subunit tRNA aminoacylation -4.28 SA1148 pheS phenylalanyl-tRNA synthetase, alpha subunit tRNA aminoacylation -4.17 SA0663 argS arginyl-tRNA synthetase tRNA aminoacylation -3.72 SA0562 lysS lysyl-tRNA synthetase tRNA aminoacylation -3.06 SA1622 glyS glycyl-tRNA synthetase tRNA aminoacylation -2.48 SA1149 pheT phenylalanyl-tRNA synthetase, beta subunit tRNA aminoacylation -1.97 SA1228 fmt methionyl-tRNA formyltransferase tRNA aminoacylation -1.80 SA1685 aspS aspartyl-tRNA synthetase tRNA aminoacylation -1.70 SA1710 valS valyl-tRNA synthetase tRNA aminoacylation -1.68 SA1686 hisS histidyl-tRNA synthetase tRNA aminoacylation -1.59 SA0574 gltX glutamyl-tRNA synthetase tRNA aminoacylation -1.50 SA1256 trmD tRNA (guanine-N1)-methyltransferase tRNA and rRNA base modification -2.90 SA1290 truB tRNA pseudouridine 55 synthase tRNA and rRNA base modification -2.65 SA1957 NA RNA methyltransferase, TrmA family tRNA and rRNA base modification -2.64 SA1694 tgt queuine tRNA-ribosyltransferase tRNA and rRNA base modification -2.29 SA1798 trmB conserved hypothetical protein TIGR00091 tRNA and rRNA base modification -1.60 SA1536 rluB pseudouridine synthase tRNA and rRNA base modification -1.57
Protein fate
SA1668 NA peptidase, U32 family Degradation of proteins, peptides, and glycopeptides -5.15 SA1667 NA peptidase, U32 family Degradation of proteins, peptides, and glycopeptides -3.70 SA1028 htrA serine protease Degradation of proteins, peptides, and glycopeptides -3.49 SA2038 NA metalloendopeptidase, putative, glycoprotease family Degradation of proteins, peptides, and glycopeptides -2.12 SA1970 sspB2 cysteine protease precursor SspB Degradation of proteins, peptides, and glycopeptides -2.09 SA1455 NA carboxyl-terminal protease Degradation of proteins, peptides, and glycopeptides -1.87 SA1036 NA protease, putative Degradation of proteins, peptides, and glycopeptides -1.81 SA1756 pepQ truncated amidase Degradation of proteins, peptides, and glycopeptides -1.72 SA2160 NA hemolysin III, putative Degradation of proteins, peptides, and glycopeptides -1.71 SA1419 NA oligoendopeptidase F, putative Degradation of proteins, peptides, and glycopeptides -1.65 SA0945 NA cytosol aminopeptidase Degradation of proteins, peptides, and glycopeptides -1.56 28
SA1692 NA protein-export membrane protein SecDF Protein and peptide secretion and trafficking -4.52 SA0969 spsB signal peptidase IB Protein and peptide secretion and trafficking -2.76 SA0968 spsA signal peptidase IA, inactive Protein and peptide secretion and trafficking -2.17 SA1253 ffh signal recognition particle protein Protein and peptide secretion and trafficking -1.93 SA1208 lspA lipoprotein signal peptidase Protein and peptide secretion and trafficking -1.65 SA1251 NA cell division protein FtsY, putative Protein and peptide secretion and trafficking -1.59 SA0844 secG preprotein translocase, SecG subunit Protein and peptide secretion and trafficking -1.58 SA1693 yajC preprotein translocase, YajC subunit Protein and peptide secretion and trafficking -1.58 SA1100 def1 polypeptide deformylase Protein modification and repair -2.87
Regulatory functions
SA0746 norR transcriptional regulator, MarR family DNA interactions -6.32 SA1107 NA transcriptional regulator, Cro/CI family DNA interactions -4.86 SA1997 NA transcriptional regulator, GntR family DNA interactions -4.43 SA1552 malR maltose operon repressor DNA interactions -3.78 SA2374 NA transcriptional regulator, TetR family, putative DNA interactions -2.96 SA0201 NA DNA-binding response regulator, AraC family DNA interactions -2.59 SA1328 glnR glutamine synthetase repressor DNA interactions -2.29 SA0249 NA transcriptional regulator, GntR family DNA interactions -2.14 SA0672 sarA staphylococcal accessory regulator A DNA interactions -1.95 SA2378 NA transcriptional regulator, AraC family DNA interactions -1.93 SA1550 NA transcriptional regulator, AraC family DNA interactions -1.87 SA2317 NA phosphosugar-binding transcriptional regulator DNA interactions -1.86 SA1535 srrA DNA-binding response regulator SrrA DNA interactions -1.80 SA2030 scrR sucrose operon repressor DNA interactions -1.51 SA2546 NA perfringolysin O regulator protein, putative Other -4.88 SA2086 NA transcriptional regulator, TenA family Other -3.69 SA1065 NA transcriptional regulator, putative Other -3.02 SA2023 agrB accessory gene regulator protein B Other -2.21 SA1398 NA transcriptional regulator, putative Other -2.19 SA0672 sarA staphylococcal accessory regulator A Other -1.95 SA2026 agrA accessory gene regulator protein A Other -1.94 SA2025 argC2 accessory gene regulator protein C Other -1.90 SA1232 NA protein kinase. POINT MUTATION Protein interactions -4.17 SA0201 NA DNA-binding response regulator, AraC family Protein interactions -2.59 SA2107 NA phosphotyrosine protein phosphatase Protein interactions -2.13 29
SA1535 srrA DNA-binding response regulator SrrA Protein interactions -1.80 SA1210 pyrR pyrimidine operon regulatory protein RNA interactions -7.54 SA2662 NA transcriptional antiterminator, BglG family RNA interactions -2.17 SA2030 scrR sucrose operon repressor Small molecule interactions -1.51
Signal transduction
SA2376 NA PTS system, sucrose-specific IIBC components, PTS -10.57 putative SA2316 NA PTS system, IIBC components PTS -8.25 SA0516 NA PTS system, IIBC components PTS -3.25 SA0175 NA PTS system, IIABC components PTS -3.21 SA2552 NA PTS system, IIABC components PTS -2.13 SA1917 NA PTS system, IIC component PTS -1.77 SA0201 NA DNA-binding response regulator, AraC family Two-component systems -2.59 SA1535 srrA DNA-binding response regulator SrrA Two-component systems -1.80
Cell envelope
SA0936 dltB DltB protein Biosynthesis and degradation of surface polysaccharides -8.60 and lipopolysaccharides SA1062 atl bifunctional autolysin Biosynthesis and degradation of surface polysaccharides -7.06 and lipopolysaccharides SA0938 dltD DltD protein Biosynthesis and degradation of surface polysaccharides -5.25 and lipopolysaccharides SA2298 NA N-acetylmuramoyl-L-alanine amidase, family 4 Biosynthesis and degradation of surface polysaccharides -3.47 and lipopolysaccharides SA2670 NA glycosyl transferase, group 1 family protein Biosynthesis and degradation of surface polysaccharides -1.94 and lipopolysaccharides SA0147 cap5L capsular polysaccharide biosynthesis protein Cap5L Biosynthesis and degradation of surface polysaccharides -1.90 and lipopolysaccharides SA0937 dltC D-alanyl carrier protein Biosynthesis and degradation of surface polysaccharides -1.85 and lipopolysaccharides SA0548 NA polysaccharide biosynthesis protein Biosynthesis and degradation of surface polysaccharides -1.74 and lipopolysaccharides SA0117 NA polysaccharide extrusion protein Biosynthesis and degradation of surface polysaccharides -1.65 and lipopolysaccharides 30
SA0141 cap5F capsular polysaccharide biosynthesis protein Cap5F Biosynthesis and degradation of surface polysaccharides -1.53 and lipopolysaccharides SA1196 murD UDP-N-acetylmuramoylalanine--D-glutamate ligase Biosynthesis of murein sacculus and peptidoglycan -4.90 SA1195 mraY phospho-N-acetylmuramoyl-pentapeptide-transferase Biosynthesis of murein sacculus and peptidoglycan -4.88 SA1066 NA fmt protein Biosynthesis of murein sacculus and peptidoglycan -3.13 SA0699 pbp4 penicillin-binding protein 4 Biosynthesis of murein sacculus and peptidoglycan -2.59 SA1329 femC glutamine synthetase FemC Biosynthesis of murein sacculus and peptidoglycan -2.44 SA2092 murAA UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 Biosynthesis of murein sacculus and peptidoglycan -2.34 SA1194 pbp1 penicillin-binding protein 1 Biosynthesis of murein sacculus and peptidoglycan -2.28 SA0693 tagA UDP-N-acetyl-D-mannosamine transferase Biosynthesis of murein sacculus and peptidoglycan -2.14 SA2352 tcaA tcaA protein Biosynthesis of murein sacculus and peptidoglycan -2.08 SA1609 pbp3 penicillin-binding protein 3 Biosynthesis of murein sacculus and peptidoglycan -1.75 SA0695 NA tagG protein, teichoic acid ABC transporter protein, Biosynthesis of murein sacculus and peptidoglycan -1.74 putative SA2151 glmM phosphoglucosamine mutase GlmM Biosynthesis of murein sacculus and peptidoglycan -1.53 SA2082 NA membrane protein, putative Other -8.74 SA2582 NA membrane protein, putative Other -4.88 SA2520 NA membrane protein, putative Other -3.89 SA2676 sasA LPXTG cell wall surface anchor family protein Other -3.29 SA0411 NA membrane protein, putative Other -2.68 SA2409 fmhA fmhA protein Other -2.37 SA2511 fnbA fibronectin-binding protein A Other -2.21 SA0255 NA membrane protein, putative Other -2.06 SA0799 NA transferrin receptor Other -1.89 SA0193 NA maltose ABC transporter, maltose-binding protein, Other -1.81 putative SA0099 sirA iron compound ABC transporter, iron compound- Other -1.81 binding protein SirA SA0203 NA iron compound ABC transporter, iron compound- Other -1.73 binding protein, putative
Cellular processes
SA2054 rpoF RNA polymerase sigma-37 factor Adaptations to atypical conditions -6.13 SA2627 betA choline dehydrogenase Adaptations to atypical conditions -2.68 SA2055 rsbW anti-sigma B factor Adaptations to atypical conditions -2.66 SA1437 NA cold shock protein, CSD family Adaptations to atypical conditions -2.65 31
SA2731 NA cold shock protein, CSD family Adaptations to atypical conditions -2.32 SA0861 NA cold shock protein, CSD family Adaptations to atypical conditions -1.87 SA2628 betB betaine aldehyde dehydrogenase Adaptations to atypical conditions -1.58 SA2075 ftsW cell division protein, FtsW/RodA/SpoVE family Cell division -2.30 SA1202 ylmF YlmF protein Cell division -1.67 SA1823 arsB arsenical pump membrane protein Detoxification -1.93 SA2291 NA staphyloxanthin biosynthesis protein Pathogenesis -13.34 SA0270 NA staphyloxanthin biosynthesis protein, putative Pathogenesis -6.30 SA2652 clfB clumping factor B Pathogenesis -5.94 SA1028 htrA serine protease Pathogenesis -3.49 SA2581 NA staphyloxanthin biosynthesis protein Pathogenesis -3.17 SA2511 fnbA fibronectin-binding protein A Pathogenesis -2.21 SA2023 agrB accessory gene regulator protein B Pathogenesis -2.21 SA1970 sspB2 cysteine protease precursor SspB Pathogenesis -2.09 SA0672 sarA staphylococcal accessory regulator A Pathogenesis -1.95 SA2026 agrA accessory gene regulator protein A Pathogenesis -1.94 SA2025 argC2 accessory gene regulator protein C Pathogenesis -1.90 SA1535 srrA DNA-binding response regulator SrrA Pathogenesis -1.80 SA1184 NA exfoliative toxin, putative Pathogenesis -1.61 SA0746 norR transcriptional regulator, MarR family Toxin production and resistance -6.32 SA0743 bacA bacitracin resistance protein Toxin production and resistance -5.98 SA0935 dltA D-alanine-activating enzyme/D-alanine-D-alanyl carrier Toxin production and resistance -4.78 protein ligase SA2437 bcr bicyclomycin resistance protein Toxin production and resistance -3.75 SA2257 NA drug transporter, putative Toxin production and resistance -3.10 SA2460 NA drug transporter, putative Toxin production and resistance -2.97 SA2409 fmhA fmhA protein Toxin production and resistance -2.37 SA1328 glnR glutamine synthetase repressor Toxin production and resistance -2.29 SA2352 tcaA tcaA protein Toxin production and resistance -2.08 SA0672 sarA staphylococcal accessory regulator A Toxin production and resistance -1.95 SA0122 NA tetracycline resistance protein, putative Toxin production and resistance -1.91 SA1809 NA hypothetical protein Toxin production and resistance -1.73 SA2347 NA drug resistance transporter, EmrB/QacA subfamily Toxin production and resistance -1.72 SA1184 NA exfoliative toxin, putative Toxin production and resistance -1.61 SA2151 glmM phosphoglucosamine mutase GlmM Toxin production and resistance -1.53
Unknown function 32
SA2293 NA NAD/NADP octopine/nopaline dehydrogenase family Enzymes of unknown specificity -6.95 protein SA1098 NA metallo-beta-lactamase family protein Enzymes of unknown specificity -5.61 SA2536 NA hydrolase, haloacid dehalogenase-like family Enzymes of unknown specificity -2.60 SA1053 NA hydrolase, alpha/beta hydrolase fold family Enzymes of unknown specificity -2.19 SA0606 NA hydrolase, haloacid dehalogenase-like family Enzymes of unknown specificity -2.17 SA1674 NA helicase, putative, RecD/TraA family Enzymes of unknown specificity -2.12 SA0596 NA aminotransferase, class II Enzymes of unknown specificity -2.03 SA2297 NA monooxygenase family protein Enzymes of unknown specificity -2.02 SA0778 NA sulfatase family protein Enzymes of unknown specificity -1.80 SA1294 NA metallo-beta-lactamase family protein Enzymes of unknown specificity -1.79 SA1654 NA hydrolase, HAD-superfamily, subfamily IIIA Enzymes of unknown specificity -1.65 SA2584 isaA immunodominant antigen A General -15.49 SA1118 typA GTP-binding protein TypA General -7.18 SA2109 NA modification methylase, HemK family General -5.21 SA0507 NA LysM domain protein General -4.46 SA0553 tilS MesJ/Ycf62 family protein General -4.15 SA0723 NA LysM domain protein General -4.09 SA0809 NA GGDEF domain protein General -3.46 SA1641 lepA GTP-binding protein LepA General -3.44 SA2328 NA abortive infection protein family General -3.44 SA1548 NA AtsA/ElaC family protein General -3.27 SA1699 NA GTP-binding protein, GTP1/OBG family General -2.32 SA2080 NA HD domain protein General -2.22 SA0790 NA integral membrane domain protein General -2.17 SA0772 NA exsB protein General -2.13 SA1683 NA HesA/MoeB/ThiF family protein General -2.07 SA1653 NA GTP-binding protein, putative General -2.03 SA1240 NA DAK2 domain protein General -2.00 SA1268 gid Gid protein General -1.90 SA2018 NA abortive infection protein family General -1.87 SA1071 NA chitinase-related protein General -1.84 SA0498 NA PAP2 family protein General -1.82 SA2666 NA N-acetylmuramoyl-L-alanine amidase domain protein General -1.81 SA0812 NA degV family protein General -1.68 SA0021 yycH yycH protein General -1.65 33
SA0276 yukA diarrheal toxin General -1.62 SA1460 NA degV family protein General -1.55 SA2108 NA Sua5/YciO/YrdC/YwlC family protein General -1.52 SA2665 NA phage infection protein, putative General -1.50
Hypothetical proteins
SA0599 NA conserved hypothetical protein Conserved -10.70 SA0711 NA conserved hypothetical protein Conserved -7.91 SA1373 NA conserved hypothetical protein Conserved -6.20 SA1993 NA conserved hypothetical protein Conserved -5.25 SA1701 NA conserved hypothetical protein Conserved -5.10 SA0721 NA conserved hypothetical protein Conserved -4.26 SA0059 NA conserved hypothetical protein Conserved -4.22 SA2373 NA conserved hypothetical protein Conserved -4.08 SA1995 NA conserved hypothetical protein Conserved -3.92 SA0565 NA conserved hypothetical protein Conserved -3.62 SA0755 NA conserved hypothetical protein Conserved -3.61 SA1297 NA conserved hypothetical protein Conserved -3.59 SA0587 NA conserved hypothetical protein Conserved -3.58 SA1017 NA conserved hypothetical protein Conserved -3.46 SA0527 NA conserved hypothetical protein Conserved -3.38 SA1991 NA conserved hypothetical protein Conserved -3.36 SA2334 NA conserved hypothetical protein Conserved -3.16 SA2528 NA conserved hypothetical protein Conserved -3.00 SA1126 NA conserved hypothetical protein Conserved -2.96 SA0913 NA conserved hypothetical protein Conserved -2.93 SA1136 NA conserved hypothetical protein Conserved -2.91 SA0067 NA conserved hypothetical protein Conserved -2.77 SA1099 NA conserved hypothetical protein Conserved -2.71 SA2102 NA conserved hypothetical protein Conserved -2.66 SA1359 NA conserved hypothetical protein Conserved -2.66 SA1903 NA conserved hypothetical protein Conserved -2.53 SA1135 NA conserved hypothetical protein Conserved -2.49 SA2134 NA conserved hypothetical protein Conserved -2.46 SA1044 NA conserved hypothetical protein Conserved -2.43 SA1842 NA accessory gene regulator B Conserved -2.21 34
SA1885 NA conserved hypothetical protein Conserved -2.18 SA1840 NA conserved hypothetical protein Conserved -2.16 SA0363 NA conserved hypothetical protein Conserved -2.10 SA1928 NA conserved hypothetical protein Conserved -2.07 SA0455 NA conserved hypothetical protein Conserved -2.06 SA2592 NA conserved hypothetical protein Conserved -2.05 SA1307 NA conserved hypothetical protein TIGR00282 Conserved -2.03 SA1664 NA conserved hypothetical protein TIGR00370 Conserved -2.03 SA1483 NA conserved hypothetical protein Conserved -2.00 SA0383 NA conserved hypothetical protein Conserved -1.99 SA1990 NA conserved hypothetical protein Conserved -1.98 SA2672 NA conserved hypothetical protein Conserved -1.98 SA1112 NA conserved hypothetical protein Conserved -1.96 SA2123 NA conserved hypothetical protein Conserved -1.95 SA1481 NA conserved hypothetical protein Conserved -1.93 SA1388 NA conserved hypothetical protein TIGR00023 Conserved -1.91 SA1799 NA conserved hypothetical protein Conserved -1.91 SA0037 NA conserved hypothetical protein Conserved -1.90 SA0710 NA conserved hypothetical protein Conserved -1.87 SA2538 NA conserved hypothetical protein Conserved -1.83 SA0629 NA conserved hypothetical protein Conserved -1.81 SA1989 NA conserved hypothetical protein Conserved -1.80 SA1617 NA conserved hypothetical protein Conserved -1.80 SA0735 NA conserved hypothetical protein Conserved -1.80 SA1947 NA conserved hypothetical protein Conserved -1.79 SA2669 NA conserved hypothetical protein Conserved -1.78 SA2346 NA conserved hypothetical protein Conserved -1.76 SA2556 NA conserved hypothetical protein Conserved -1.74 SA1444 NA conserved hypothetical protein Conserved -1.72 SA0752 NA conserved hypothetical protein Conserved -1.72 SA1968 NA conserved hypothetical protein Conserved -1.71 SA1836 NA conserved hypothetical protein Conserved -1.70 SA0662 NA conserved hypothetical protein Conserved -1.67 SA1810 NA conserved hypothetical protein TIGR01212 Conserved -1.63 SA0537 NA conserved hypothetical protein Conserved -1.60 SA2371 NA conserved hypothetical protein Conserved -1.58 SA0802 NA conserved hypothetical protein Conserved -1.57 35
SA0804 NA conserved hypothetical protein Conserved -1.57 SA0271 NA conserved hypothetical protein Conserved -1.57 SA0511 NA conserved hypothetical protein Conserved -1.55 SA0529 NA conserved hypothetical protein Conserved -1.55 SA2377 NA conserved hypothetical protein Conserved -1.52 SA0580 NA conserved hypothetical protein Conserved -1.52 SA1380 NA conserved hypothetical protein Conserved -1.52 SA2557 NA conserved domain protein Domain -5.53 SA2037 NA conserved domain protein Domain -4.04
a Fold value refers to expression increases or decreases for up- or down-regulated genes respectively.