Section 1
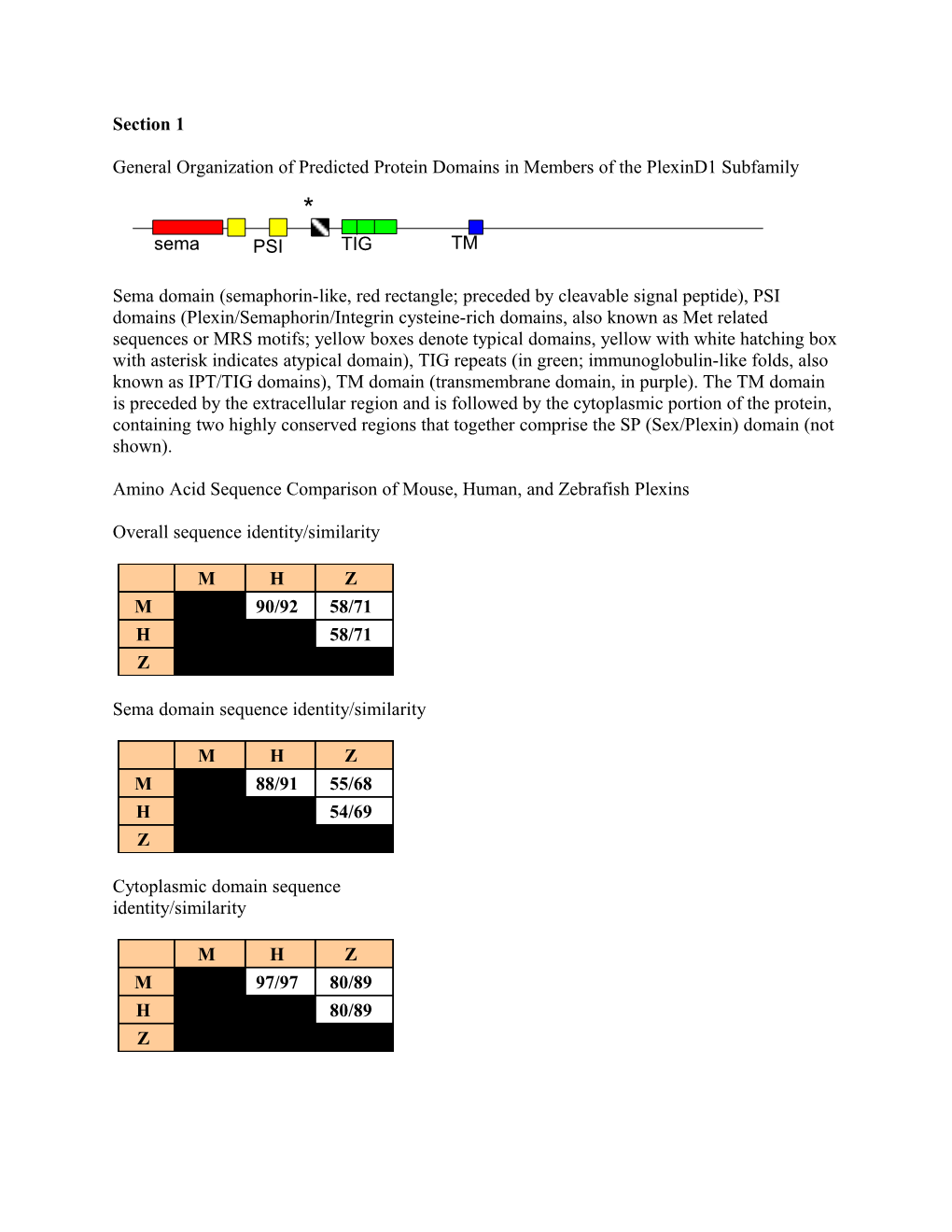
General Organization of Predicted Protein Domains in Members of the PlexinD1 Subfamily * sema PSI TIG TM
Sema domain (semaphorin-like, red rectangle; preceded by cleavable signal peptide), PSI domains (Plexin/Semaphorin/Integrin cysteine-rich domains, also known as Met related sequences or MRS motifs; yellow boxes denote typical domains, yellow with white hatching box with asterisk indicates atypical domain), TIG repeats (in green; immunoglobulin-like folds, also known as IPT/TIG domains), TM domain (transmembrane domain, in purple). The TM domain is preceded by the extracellular region and is followed by the cytoplasmic portion of the protein, containing two highly conserved regions that together comprise the SP (Sex/Plexin) domain (not shown).
Amino Acid Sequence Comparison of Mouse, Human, and Zebrafish Plexins
Overall sequence identity/similarity
M H Z M 90/92 58/71 H 58/71 Z
Sema domain sequence identity/similarity
M H Z M 88/91 55/68 H 54/69 Z
Cytoplasmic domain sequence identity/similarity
M H Z M 97/97 80/89 H 80/89 Z Radiation Hybrid Mapping of Zebrafish plxnD1
Radiation hybrid mapping on the Goodfellow T51 radiation hybrid panel using the plxnD1 intron primer 5-TCCCGACTACAGCACGCACCAATA-3 and exon primer 5- GCCGCCGCAGCTCGCAGAAG-3 located this gene to the zebrafish LG8 bottom telomere (marker “unp2202#” in the Tübingen Map of the Zebrafish Genome: http://wwwmap.tuebingen.mpg.de/).
Section 2
Morpholino Oligonucleotides
Morpholino oligonucleotides (morpholinos) used in this work were purchased from Gene Tools, LLC. Morpholino sequences and specificities were as follows:
MORPHOLINO SEQUENCE Control (general negative control, not gene specific) 5-CCTCTTACCTCAGTTACAATTTATA- 3 plxnD1 2215-2304 (to prevent proper splicing of the exon encoded 5-TGAGGGTATTTACAGTCGCTCCGC-3 by bases 2215-2304 in the plxnD1 cDNA ORF) plxnD1 2215-2304 negative control 5-TcAGGcTATTTAgAGTCcCTCCcC-3 (4-base mismatch control for the above morpholino) plxnD1 3207-3462 (to prevent correct splicing of the exon encoded 5- by bases 3207-3462 in the plxnD1 cDNA ORF) CACACACACTCACGTTGATGATGAG-3 plxnD1 3207-3462 negative control 5-CAgACAgACTCACcTTGATcATcAG-3 (4-base mismatch control for the above morpholino) sema3A15’UTR (to block translation of the sema3A1 mRNA 5- message, complementary to bases –33 to –60 of the sema3A1 GCTTGTAGCCCACAGTGCCCAGAGC-3 5UTR; 12) sema3A15’UTR negative control 5-CTTcTAGCCgACAGaGCCCAGtGCA-3 (mismatch control for the above morpholino) sema3A2550-670 (to prevent appropriate splicing of the exon 5-AAATGTGTCTTACCGTTGAGCCATC- encoded by bases 550-670 in the sema3A2 cDNA ORF) 3 sema3A2550-670 negative control 5-AAATcTcTCTTAgCGTTcAGCgATC-3 (4-base mismatch control for the above morpholino)
The position of the mentioned exons in the cDNA sequence is defined by considering the first base of the ORF as +1. For the negative control morpholinos, cognate mismatched bases are shown in lower case.
Primer Pairs for Assaying plxnD1 Morpholino Effects
Primers used to determine the specificity and efficiency of the plxnD1 splicing blocking morpholinos: GENE TARGETED PRIMER PAIR SEQUENCES EXPECTED EXON PRODUCT SIZE plxnD1 2215-2304 5-GGATCTCCACCAACGCCACTGTTCACC-3 669 bp 5-CACACTCCTGACCGAGCGCTGCACG-3 plxnD1 3207-3462 5-AGCGCTTCACCAAACTTCACATTCAGCTA- 547 bp 3 5-TGGCACTCAGTGATCTGTTGATGC-3 plxnD1 3207-3462 5-AGCGCTTCACCAAACTTCACATTCAGCTA- 933 bp 3 5-ACGCCTCTCATAGTCACAGCACATC-3 -actin N/A 5-CAGCTAGTGCGAATATCATCT-3 ~200 bp 5-TTTCTGTCCCATACCAACC-3
To determine the specificity and activity of the morpholinos designed to block the splicing of particular plxnD1 exons, we isolated total mRNA from un-treated embryos, control and plxnD1 morphants at 28–32 hpf (morphants made with 4.6 ng of the corresponding morpholino) and performed reverse transcription-PCR (RT-PCR) with primer pairs designed to amplify cDNA fragments containing the plxnD1 exons targeted in the plxnD1 morphants. Aberrant splicing of the targeted plxnD1 exons was observed only in the corresponding plxnD1 morphants, but not in either control or in the nonequivalent plxnD1 morphant. In addition, the amounts of properly spliced plxnD1 mRNA were significantly reduced in both plxnD1 morphants, with the reduction being more pronounced in plxnD13207-3462 morphants (see below), correlating with their somewhat stronger vascular guidance phenotype (data not shown).
Normal Splicing of plxnD1 mRNA Is Substantially Reduced in plxnD1 Morphants
In red: DNA molecular weight markers “t” (1 Kb ladder) and “h” (100 bp). RT-PCR reactions products from untreated embryos (1), control morphants (2), plxnD12215-2304 morphants (3), and plxnD13207-3462 morphants (4).
In blue: Legend refers to the exon or cDNA fragment being amplified from the indicated gene by RT-PCR. For the plxnD13207-3462 morphants, two different sets of primer pairs were used for the RT-PCR reactions, expected to yield a band of 547 bp (plxnD1 3207-3462A) or a product of 933 bp (plxnD1 3207-3462B). Asterisks mark the RT-PCR reactions showing aberrant splicing. Yellow arrowheads point to the band of RT-PCR product corresponding to normal plxnD1 mRNA splicing reactions in the plxnD1 morphants.
Specificity and Efficiency of the sema3A1 Morpholino
The inhibitory effect on translation of sema3A1 mRNA by the sema3A15’UTR morpholino has been previously established (Shoji et al., 2003) using an almost identical morpholino and was therefore not assayed.
Section 3 Injection of plxnD1 Morpholinos into obd Mutants
To further test the identity between plxnD1 and obd, we injected the plxnD12215-2304 morpholino into embryos derived from mating obdfov01b homozygous mutants with heterozygous obdfs31 fish carrying an endothelial-specific GFP reporter. The vascular phenotype of plxnD1 morphants and double obd mutants-plxnD1 morphants is undistinguishable from that of obd mutants. The lack of enhancement of the obd phenotype by the plxnD1 morpholino further supports the idea that these obd alleles represent loss of function mutations in the plxnD1 gene.
Rescue of obd Mutants
Demonstrating phenotypic rescue of a mutant by the corresponding wild type gene is desirable as evidence for identification of the correct gene, and we attempted to rescue obd mutants by injection of either in vitro synthesized plxnD1 mRNA or DNA expression constructs that use the CMV promoter to drive plxnD1 overexpression. Both of these methods have serious technical limitations for mutants like obd (exhibiting late phenotypes) and these attempts were not successful. Injected mRNA is rapidly degraded within zebrafish embryos and the obd phenotype appears relatively late (at approximately 1 day postfertilization), making it unlikely that any injected plxnD1 transcript remains intact when its function is normally required. Injected DNA expression constructs can deliver mRNA at later stages of development but expression is mosaic with only a small fraction of cells in the animal showing expression. Our currently available tools and methods do not allow us to score rescue in a few isolated cells. Future rescue attempts will use transgenic tools to drive tissue-specific gene expression in a nonmosaic manner.