Bacterial melanin interacts with double- stranded DNA with high affinity and may inhibit cell metabolism in vivo
Jing Geng · Peng Yuan · Chun Shao · Sheng-Bing Yu · Bo Zhou · Ping Zhou · Xiang- Dong Chen*
Supplementary Materials
UV-visible absorbance spectroscopy
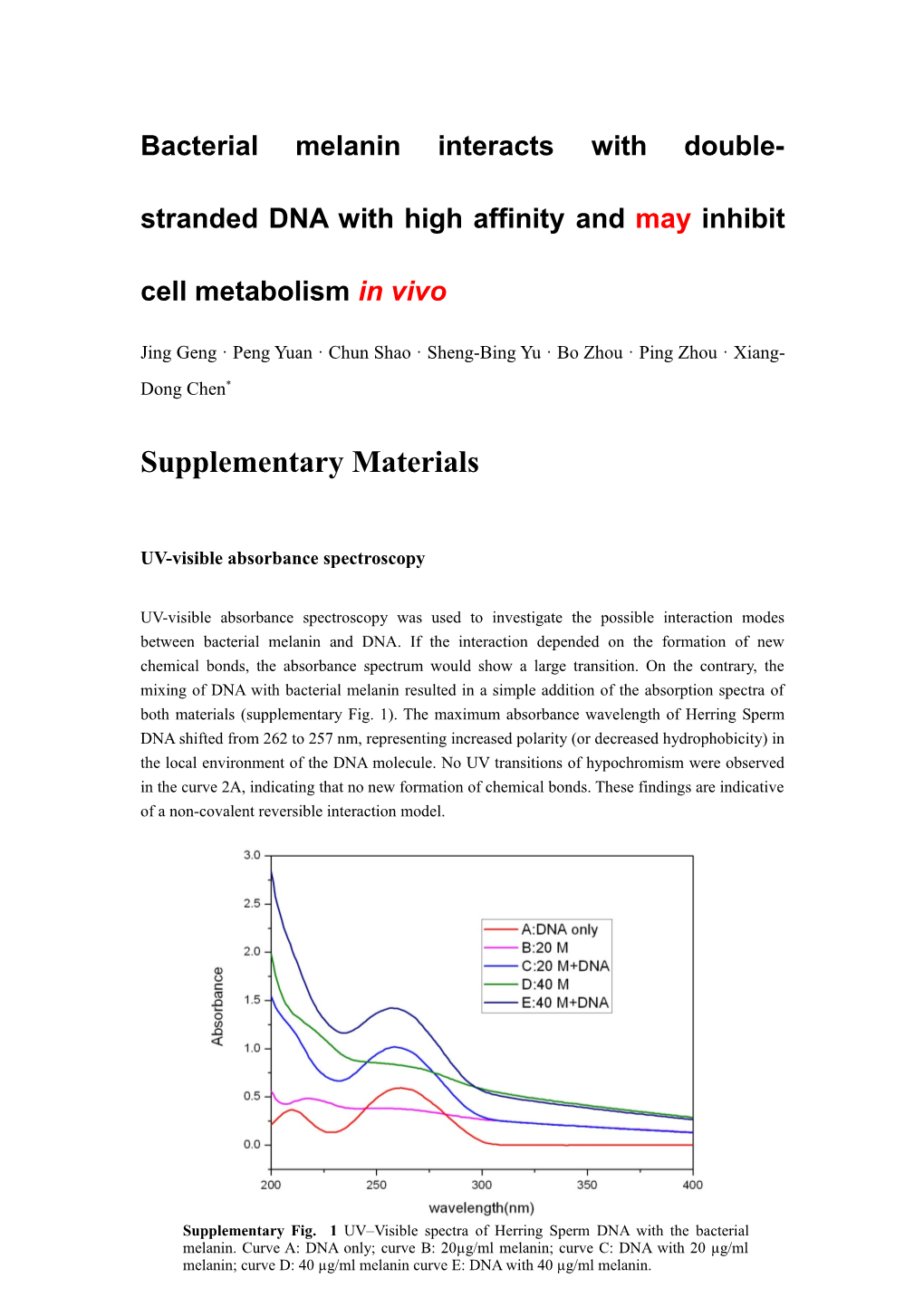
UV-visible absorbance spectroscopy was used to investigate the possible interaction modes between bacterial melanin and DNA. If the interaction depended on the formation of new chemical bonds, the absorbance spectrum would show a large transition. On the contrary, the mixing of DNA with bacterial melanin resulted in a simple addition of the absorption spectra of both materials (supplementary Fig. 1). The maximum absorbance wavelength of Herring Sperm DNA shifted from 262 to 257 nm, representing increased polarity (or decreased hydrophobicity) in the local environment of the DNA molecule. No UV transitions of hypochromism were observed in the curve 2A, indicating that no new formation of chemical bonds. These findings are indicative of a non-covalent reversible interaction model.
Supplementary Fig. 1 UV–Visible spectra of Herring Sperm DNA with the bacterial melanin. Curve A: DNA only; curve B: 20µg/ml melanin; curve C: DNA with 20 µg/ml melanin; curve D: 40 µg/ml melanin curve E: DNA with 40 µg/ml melanin. Electrospray ionization mass spectrometry
Different melanin polymers could be distinguished by their molecular mass with electrospray ionization mass spectrometry (ESI-MS). Although this analysis does not reveal the exact structure of the melanin, MS analysis has been frequently employed for understanding the subunit composition of melanin (Allegri et al., 1996, Bertazzo et al., 1999). The monomers were oxidized from tyrosine with some chemical modification (Kaxiras et al., 2006, Simon et al., 2004). Napolitano et al. (1996) recognized a consensus m/z difference patterns at 137 and 206 corresponded to DHI and ‘DHI-like’ groups (such as DHICA, DOPA). This m/z patterns indicate how many residues of oxidized tyrosine are added to the oligomer. In the present study, ESI-MS analysis of the bacterial melanin found it was composed of a series of small oligomers with an m/z ranging from 100~580. The same MS pattern was identified in our present ESI-MS analysis for bacterial melanin, with m/z identifier series located at: 99.8, 231.8, 367,522; 141.5, 284.8, 487.8; 178.7, 310.8, 443.4, 573.8 and 190.8, 324.7, 522, indicating that melanin may be comprised of DHI, DHICA and DOPA oligomers (supplementary Fig. 2). The majority of the m/z pattern signals were located in the 200 to 400 range, indicating an abundance of bacterial melanin dimers, while it is possible that monomers (m/v<200) and a few possible tetramers (443.4, 487.8, 522 and 573.8) may also exist.
070618-3 #99 RT: 1.56 AV: 1 NL: 8.48E4 T: - c m s [ 50.00-800.00] 155.1 100
95
90
85
80 211.8 75 324.7
70
65 e
c 60 n a
d 190.8 n 55 212.9 u b A
e 50 v i t a l
e 45 R
40 213.8 270.3
35 310.9 141.2 284.8 30 231.6
25 175.7 256.6 367.0
20 497.8 573.0 15 522.0 10 382.2 572.1 99.6 128.2 443.4 5
0 50 100 150 200 250 300 350 400 450 500 550 600 650 700 750 800 m/z Supplementary Fig. 2 EMI-MS spectrum of bacterial melanin and different structures of melanin. Most signals were distributed in dimeric range Supplementary references
Allegri, G., Bertazzo, A., Costa, C., Seraglia, R.and 'Kaldi, P. (1996) Investigation on melanin biosynthesis from 5,6-dihydroxytryptamine by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom, 10, 419-423. Bertazzo, A., Costa, C.V.L., Allegri, G., Schiavolin, M., Favretto, D.and Traldi, P. (1999) Enzymatic oligomerization of tyrosine by tyrosinase and peroxidase studied by matrixassisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom, 13, 542-547. Kaxiras, E., Tsolakidis, A., Zonios, G.and Meng, S. (2006) Structural model of eumelanin. Phys Rev Lett, 97, 218102. Napolitano, A., Pezzella, A.and Prota, G. (1996) Structural analysis of synthetic melanins from 5,6- dihydroxyindole by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom, 10, 468-472. Simon, J.D.and Ito, S. (2004) The Chemical Structure of Melanin. Pigment Cell Res, 17, 423-424.