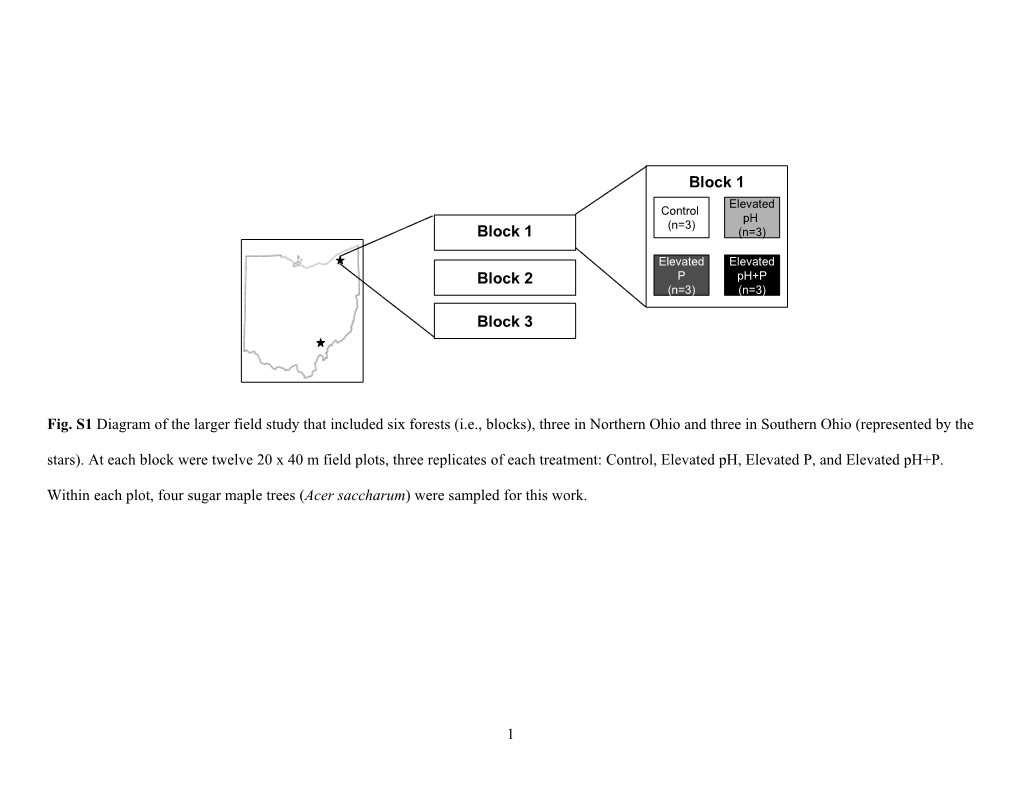
Block 1 Elevated Control pH (n=3) Block 1 (n=3)
Elevated Elevated Block 2 P pH+P (n=3) (n=3)
Block 3
Fig. S1 Diagram of the larger field study that included six forests (i.e., blocks), three in Northern Ohio and three in Southern Ohio (represented by the stars). At each block were twelve 20 x 40 m field plots, three replicates of each treatment: Control, Elevated pH, Elevated P, and Elevated pH+P.
Within each plot, four sugar maple trees (Acer saccharum) were sampled for this work.
1 P7mF P7mR
P3F P4F P6F P6R P4R P3R
P6RFor P4R
179 bp fragment
Fig. S2 Diagram of the relative locations of the phosphate transporter (PT) primers from
Sokolski et al. (2011). The primers used in the current study were P6R in the opposite direction (called “P6RFor”) as the forward primer and P4R as the reverse primer. The primers used are shown in gray. The overall aim of the study was to use quantitative
PCR, thus primers amplifying a small DNA fragment (179 bp) were selected. The primers yielded a DNA fragment that matched with AM fungal-specific PT genes (Tables
1, S1, and S4).
2 Table S1. Primer sets tested in the initial optimization of the experiment with the ultimate goal of amplifying PT genes from environmental samples.
The primer sets were used to amplify PT genes from inocula of Rhizophagus irregularis, Funneliformis mosseae, and Acaulospora sp.
Forward Primer and reference Primer Sequence Reverse Primer and reference Primer Sequence Result P4F GAATTRATGATYATYATTGTYG P6Fa AAATCCTTGCATAGCAAATACT No amplified (Sokolski et al. 2011) C (Sokolski et al. 2011) fragments P4F GAATTRATGATYATYATTGTYG P7mFa GAATCCCTGCATCGCGAATAC No amplified (Sokolski et al. 2011) C (Sokolski et al. 2011) fragments P4F GAATTRATGATYATYATTGTYG InGiPTFa,b GCCAACAATAATCAACAGCAG No amplified (Sokolski et al. 2011) C (Maldonado-Mendoza et al. fragments 2001) P4F GAATTRATGATYATYATTGTYG PT2Aa TAACATCATCTTGAATACGATTC No amplified (Sokolski et al. 2011) C (Benedetto et al. 2005) fragments GvPTFc TGGTCATGGTATATCAGCTG P4R AYYCTTTCTTTCCTTTCTTCAACG No amplified (Harrison and van Buuren 1995) (Sokolski et al. 2011) fragments P6Ra AAACTAAAAGACATTGGTGGAC P4R AYYCTTTCTTTCCTTTCTTCAACG Fragment of (Sokolski et al. 2011) (Sokolski et al. 2011) correct size for R. irregularis innoculum aThese primers were designed as the reverse compliment of what is reported in the reference in order to have an amplicon of a size suitable for quantitative PCR. bThe primer from Maldonado-Mendoza et al. (2001) was unnamed in the paper, but is the forward primer from the internal fragment used to create their probe. cThe primer from Harrison and van Buuren (1995) was unnamed in the paper, but is primer number 1 for amplifying phosphate transporter gene from
Funneliformis mosseae.
3 M C L C X L X L X P P C P + B
2000bp 1200bp 800bp 400bp 200bp 100bp
Fig. S3 Gel picture of PCR using DNA extracted from cleaned and lyophilized roots (i.e., preliminary samples; see text for details) as template and PT-specific primers, P6RFor and P4R. Lane headings are as follows: M – Low mass ladder (Life Technologies
Corporation, Carlsbad, CA, USA); C and P – root samples from a plot that did not receive lime (C - Control or P - elevated P plots); L and X – root samples from a plot that received lime (L - elevated pH or X - elevated pH+P plots); + – PCR positive control using DNA extracted from Rhizophagus irregularis spores, B – PCR blank.
4 Plot 1 Plot 7
M DNA DNA RNA cDNA DNA DNA RNA cDNA M 2000bp 1200bp 800bp
400bp
200bp
100bp
Fig. S4 Gel of PCR products using the extracts from RNA isolation and reverse transcription as template and PT-specific primers, P6RFor and P4R on two uncleaned root samples. Lane headings are as follows: M – Low mass ladder (Life Technologies
Corporation, Carlsbad, CA, USA); DNA – PCR product using extracted nucleic acids as template; RNA – PCR product using DNase-treated nucleic acids as template; cDNA –
PCR product using reverse-transcribed total RNA as template.
5 10 20 30 40 50 60 70 80 90 100 110 120 130
....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|....|... Pr-HG380725 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380726 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380727 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380728 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380729 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380730 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380731 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380732 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380733 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCAYGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380734 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAARAATTAGCAAACGAAGACCATGATTACAA Pr-HG380735 CAAACGCATTTGTTGGTCCATTGCTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCCCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380736 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACGA Pr-HG380737 CAAACGCATTTGTTGGACCATTACTTTTAATCTCTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380738 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380739 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380940 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380741 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380742 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380743 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATCAGCAAACGAAGACCATGATTACAA Pr-HG380744 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380745 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380746 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA Pr-HG380747 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAGACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGGAGACCATGATTACAA Pr-HG380748 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380679 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380681 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380682 CAAACGCATTTGTTAGACCATTACTTTTAATCTTTGCCGCATGGATGTTTACTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAGGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380684 CAAACGCATTTGTTGGACCATTACTATTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380685 CAAACGCTTTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC–HG380686 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATT~~~~~~~~~~~~~~~~~~~~~~~~ TC-HG380687 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGAGCATGATTACAA TC-HG380688 CAAACGCATTTGCTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380689 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380690 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380691 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380692 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACA~ TC-HG380693 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTAGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380694 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380695 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380696 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCA~~~~~~~~~ TC-HG380697 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380698 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380699 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGATGAATTAGCAAACGAAGACCATGATTACAA TC-HG380700 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~
6 TC-HG380701 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380702 CAAACGCATTTGTTGAACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380703 ~~~~~~~~~TTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380704 ~~~~~GCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380705 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCTCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGA~~~~~~ TC-HG380706 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380707 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGA~~~~~~ TC-HG380708 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380709 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCA~~~~~~~~~ TC-HG380710 ~~~~~GCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380711 ~~~~~GCATTTGTTGGACCATTACTTTWAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380712 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCACGGATGTTTATTGGTGGTATATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380713 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTTCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGA~~~~~~ TC-HG380714 CAAACGCATTTGTWGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGA~~~~~~ TC-HG380716 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATATTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380718 ~~~~~GCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCATGATAACAA TC-HG380719 ~~~~~~~~~~~~~~GGTCCATTAATTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTATACTCTCATTTCTTATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTAGCAAACGAAGACCATGATTACAA TC-HG380720 CAAACGCATTTGTTGGTCCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTWTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTRTCCCTTGAAGAATTAGCAAACGAAGACCATGATT~~~~ TC-HG380721 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACCAT~~~~~~~~ TC-HG380722 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTTATTCCTGAAACAAAAGGTTTATCCCTTGAAGAATTAGCAAACGAAGACC~~~~~~~~~~ TC-HG380723 CAAACGCATTTGTTGGACCATTACTTTTAATCTTTGCCGCATGGATGTTTATTGGTGGTTTATTCTCATTCCTAATTCCTGAAACAAAAGGTTTGTCCCTTGAAGAATTACGAAA~~~~~~~~~~~~~~~~~~ AF359112 CAAACGCATTTGTTGGACAATTGCTTTTAATCTTTGCCGCATGGATGTTTATTGGGGGTTTATTCTCAATACTCATTCCTGAAACAAAAGGTTTGTCTCTTGAAGAATTAGCAAATGAAGACCATGATTTCAA AY037894 CAAACGCATTTGTTGGACAATTGCTTTTAATCTTTGCCGCATGGATGTTTATTGGGGGTTTATTCTCAATACTCATTCCTGAAACAAAAGGTTTGTCTCTTGAAGAATTAGCAAATGAAGACCATGATTTCAA U38650 CAAACGCATTTGTAGGACCATTACTTTTAATCTTTTCCGCATGGATGTTTATTGGCGGTTTATTCTCAATACTTATTCCTGAAACAAAAGGTTTATCACTTGAAGAATTAGCCAACGAAGAACATACTTATGA DQ074452 CAAACGCATTTGTTGGACAATTACTTTTAATCTTTTCCGCATGGATGTTTATTGGGGGTTTATTCTCAATATTAATTCCTGAAACAAAAGGTTTGTCTCTTGAAGAATTAGCAAACGAAGACCATGAATACAA HQ896039 CAAACGCATTTGTTGGACAATTGCTTTTAATCTTTGCCGCATGGATGTTTATTGGGGGTTTATTCTCAATACTCATTCCTGAAACAAAAGGTTTGTCTCTTGAAGAATTAGCAAATGAAGACCATGATTTCAA
Fig. S5 Sequence alignment of the 24 preliminary sample sequences (cloned from genomic DNA; clones are named with “Pr” followed by the sequence accession number), the 41 time course sample sequences (cloned from cDNA; clones are named with “TC” followed by the sequence accession number), and five AM fungal-specific PT sequences that match with our amplified fragment (listed by their accession numbers: AF359112
– Rhizophagus irregularis complete cds (Maldonado-Mendoza et al. 2001); AY037894 – R. irregularis partial cds (Maldonado-Mendoza et al.
2001); GVU38650 – Glomus versiforme complete cds (Harrison and van Buuren 1995); DQ074452 – Funneliformis mosseae partial cds (Benedetto et al. 2005); HQ896039 – Rhisophagus irregularis partial cds (unpublished)).
7 8 M
21,226bp
564bp
Fig. S6 Gel of extracted nucleic acids from a subset of uncleaned root samples that were preserved in liquid nitrogen in the field and stored at -70°C until extraction (i.e., time course samples; 48 total samples were collected; see text for details). All 48 samples were run in the agarose gels after three years of storage at -20°C and still showed a large quantity of high molecular weight DNA and bands of 18S and 28S RNA. This indicates good purity of the extracted nucleic acids. Lane 1 (marked with “M”) contains the lambda DNA/EcoRI + HindIII marker (Thermo Fisher Scientific Inc., Waltham, MA,
USA).
9 M S + B
2000bp
1200bp 800bp
400bp
200bp
Fig. S7 Gel showing a subset of PCR products from cDNA spiked with 25 copies of plasmid DNA containing the PT gene as template. A total of 39 spiked cDNA samples were PCR-amplified and all produced a fragment of the expected size (179 bp). Positive
PCR reactions for all spiked samples indicate that there are no inhibitory effects of soil extracts. Lane headings are as follows: M – Low mass ladder (Life Technologies
Corporation, Carlsbad, CA, USA); S– positive control using cDNA from a root sample that worked previously also spiked with 25 copies/μl of plasmid containing the PT gene;
+ – PCR positive control using a plasmid containing the PT gene; B – PCR blank.
10 Table S2 Average (± standard error) Pi (determined with sodium bicarbonate extraction) in bulk soil from the treatment plots (n= 3 plots per treatment) collected with the time course samples and the two way analysis of variancea table showing the effects of treatment and date on the Pi measurments.
Treatment 23 May 2011 1 June 2011 8 June 2011 22 June 2011 All Datesa Control 10.74 (±0.77) 9.76 (±1.40) 12.82 (±1.58) 10.99 (±0.75) 11.08 (±0.61)a Elevated pH 9.96 (±0.79) 10.56 (±1.08) 9.77 (±0.79) 10.11 (±1.58) 10.10 (±0.48)a Elevated P 28.24 (±7.89) 35.20 (±9.78) 38.69 (±19.93) 35.48 (±11.90) 34.40 (±5.74)b Elevated pH+P 17.87 (±3.50) 37.22 (±2.83) 31.73 (±1.82) 29.65 (±9.73) 29.12 (±3.15)b ANOVA Table Source of Variation DF SS MS F P Treatment 3 5552.521 1850.840 11.960 <0.001 Date 3 342.195 114.065 0.737 0.538 Treatment x Date 9 446.009 49.557 0.320 0.962 Residual 32 4951.962 154.749 Total 47 11292.688 240.270 aDifferent superscript letters indicate significant differences (α=0.05) between treatments as determined with a two way ANOVA conducted in SigmaStat, version 3.5 (Systat Software, Inc., 2006) using treatment and date as factors and the Holm-Sidak multiple comparison test. Date was not a significant factor.
11 Table S3 Average (± standard error) pH measured in bulk soil from the treatment plots (n= 3 plots per treatment) collected with the time course samples and the two way analysis of variancea table showing the effects of treatment and date on the soil pH.
Treatment 23 May 2011 1 June 2011 8 June 2011 22 June 2011 All Datesa Control 4.21 (±0.090) 4.27 (±0.075) 4.02 (±0.046) 4.07 (±0.043) 4.14 (±0.042)a Elevated pH 5.70 (±0.32) 5.96 (±0.051) 6.32 (±0.24) 5.79 (±0.37) 5.94 (±0.14) b Elevated P 4.26 (±0.074) 4.22 (±0.07) 4.09 (±0.012) 4.05 (±0.063) 4.16 (±0.037) a Elevated pH+P 6.16 (±0.17) 6.18 (±0.19) 5.74 (±0.27) 6.01 (±0.19) 6.02 (±0.10) b ANOVA Table Source of Variation DF SS MS F P Treatment 3 40.318 13.439 141.905 <0.001 Date 3 0.197 0.0657 0.694 0.563 Treatment x Date 9 1.055 0.117 1.238 0.308 Residual 32 3.031 0.0947 Total 47 44.600 0.949 aDifferent superscript letters indicate significant differences (α=0.05) between treatments as determined with a two way ANOVA conducted in SigmaStat, version 3.5 (Systat Software, Inc., 2006) using treatment and date as factors and the Holm-Sidak multiple comparison test. Date was not a significant factor.
12 TABLE S4 Average (± standard error) AM fungal 18S gene copy number (gene copies (g dry root)-1) in uncleaned root samples from the treatment plots (n= 3 plots per treatment) collected with the time course samples and two way analysis of variancea table testing the effects of treatment and date on 18S gene expression.
Treatment 23 May 2011 1 June 2011 8 June 2011 22 June 2011 All Datesa Control 1.8x107 (±0.3x107) 2.3x107 (±1.4x107) 2.8x107 (±1.0x107) 1.3x107 (±0.8x107) 2.0x107 (±0.4x107) Elevated pH 1.9x107 (±0.6x107) 4.9x107 (±4.2x107) 3.2x107 (±1.4x107) 1.8x107 (±0.8x107) 3.0x107 (±1.0x107) Elevated P 2.4x107 (±1.0x107) 1.0x107 (±0.5x107) 8.0x107 (±7.0x107) 4.0x107 (±1.4x107) 3.9x107 (±1.7x107) Elevated pH+P 2.9x107 (±0.8x107) 3.3x107 (±1.4x107) 2.9x107 (±1.5x107) 4.8x107 (±2.8x107) 3.5x107 (±0.8x107) ANOVA Table Source of Variation DF SS MS F P Treatment 3 2.26x1015 7.54 x1014 0.451 0.718 Date 3 2.52 x1015 8.40 x1014 0.502 0.683 Treatment x Date 9 8.87 x1015 9.85 x1014 0.589 0.796 Residual 32 5.35 x1016 1.67 x1015 Total 47 6.72 x1016 1.43 x1015 aTwo way ANOVA conducted in SigmaStat, version 3.5 (Systat Software, Inc., 2006) using treatment and date as factors and the
Holm-Sidak multiple comparison test indicate that treatment and date were not significant factors.
13 References
Benedetto A, Magurno F, Bonfante P, Lanfranco L (2005) Expression profiles of a phosphate transporter gene (GmosPT) from the endomycorrhizal fungus Glomus mosseae. Mycorrhiza 15:620–627
Harrison MJ, van Buuren ML (1995) A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378:626–629
Maldonado-Mendoza IE, Dewbre GR, Harrison MJ (2001) A phosphate transporter gene from the extra-radical mycelium of an arbuscular mycorrhizal fungus Glomus intraradices is regulated in response to phosphate in the environment. Mol Plant Microbe Interact 14:1140–1148
Sokolski S, Dalpé Y, Piché Y (2011) Phosphate transporter genes as reliable gene markers for the identification and discrimination of arbuscular mycorrhizal fungi in the genus Glomus. Appl Environ Microbiol 77:1888–1891
14