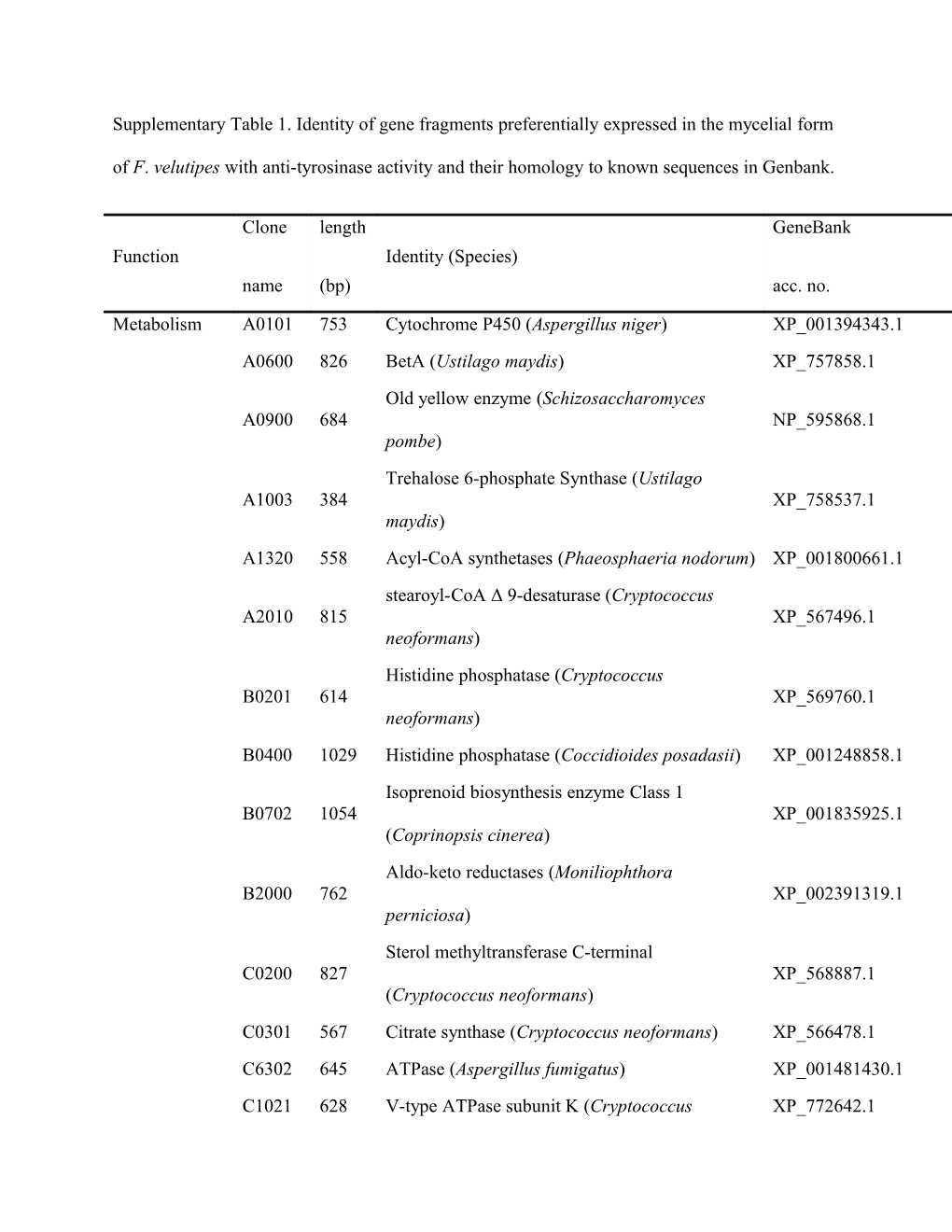
Supplementary Table 1. Identity of gene fragments preferentially expressed in the mycelial form of F. velutipes with anti-tyrosinase activity and their homology to known sequences in Genbank.
Clone length GeneBank Function Identity (Species) name (bp) acc. no.
Metabolism A0101 753 Cytochrome P450 (Aspergillus niger) XP_001394343.1 A0600 826 BetA (Ustilago maydis) XP_757858.1 Old yellow enzyme (Schizosaccharomyces A0900 684 NP_595868.1 pombe) Trehalose 6-phosphate Synthase (Ustilago A1003 384 XP_758537.1 maydis) A1320 558 Acyl-CoA synthetases (Phaeosphaeria nodorum) XP_001800661.1
stearoyl-CoA Δ 9-desaturase (Cryptococcus A2010 815 XP_567496.1 neoformans) Histidine phosphatase (Cryptococcus B0201 614 XP_569760.1 neoformans) B0400 1029 Histidine phosphatase (Coccidioides posadasii) XP_001248858.1 Isoprenoid biosynthesis enzyme Class 1 B0702 1054 XP_001835925.1 (Coprinopsis cinerea) Aldo-keto reductases (Moniliophthora B2000 762 XP_002391319.1 perniciosa) Sterol methyltransferase C-terminal C0200 827 XP_568887.1 (Cryptococcus neoformans) C0301 567 Citrate synthase (Cryptococcus neoformans) XP_566478.1 C6302 645 ATPase (Aspergillus fumigatus) XP_001481430.1 C1021 628 V-type ATPase subunit K (Cryptococcus XP_772642.1 neoformans) Lactate dehydrogenase (Cryptococcus C1022 983 XP_571419.1 neoformans) C1401 521 Catalase (Cryptococcus neoformans) XP_568133.1 C1500 941 NOX/Duox family (Aspergillus niger) XP_001393108.1 C1701 926 Enolase (Ustilago maydis) XP_759503.1 D0703 546 TPP-binding module (Cryptococcus neoformans) XP_570402.1 D0800 367 Thioesterase (Aspergillus fumigatus ) XP_752445 D1111 428 Coenzyme A pyrophosphatase (Ustilago maydis) XP_759099.1 D1121 452 Glycogen synthase (Cryptococcus neoformans) XP_572121.1 D1131 350 GMC oxidoreductase (Aspergillus niger) XP_001398998.1 Peptidase C19 containing ubiquitinyl hydrolase D1140 292 XP_758487.1 (Ustilago maydis) Zn-dependent alcohol dehydrogenase (Ustilago D1301 680 XP_758131.1 maydis) Sulfite reductase subunit β (Cryptococcus D1401 417 XP_773519.1 neoformans) E0102 351 Copper binding protein (Aspergillus fumigatus) XP_754260.1 N-Acetyl glutamate kinase (Vanderwaltozyma E0202 453 XP_001642397.1 polyspora) Glyceraldehyde-3-phosphate dehydrogenase E1900 965 XP_758638.1 (Ustilago maydis) Mannitol-1-phosphate dehydrogenase F0821 237 XP_571772.1 (Cryptococcus neoformans) F1901 688 Catalase (Phaeosphaeria nodorum ) XP_001805033.1 Gene A0102 383 Ribosomal protein S27 (Ustilago maydis) XP_757782.1 expression and A0103 291 GAL4 (Aspergillus nidulans) XP_659726.1 signal Src homology 3 domains (Cryptococcus A0302 265 XP_569072.1 transduction neoformans) A0901 590 Ras-related nuclear proteins (Ustilago maydis) XP_757511.1 Forkhead associated domain (Phaeosphaeria B0102 430 XP_001792134.1 nodorum) C2022 319 Nedd8 (Aspergillus nidulans) XP_663783.1 D7101 480 Ras-like small GTPases (Pichia pastoris) XP_002493629.1 D7022 426 Ras-like proteins (Pichia pastoris) XP_002493629.1 E0101 416 Methionyl-tRNA synthetase (Pichia stipitis) XP_001382405.1 Cutinase transcription factor 1 β (Gibberella E0201 275 XP_387368.1 zeae) Fungal specific transcription factor domain E0302 632 XP_760639.1 (Ustilago maydis) E0401 922 DnaJ-like protein (Candida albicans) XP_712644.1 F0201 441 Peroxiredoxin family (Yarrowia lipolytica) XP_500241.1 Translational cofactor EF-1 γ; Cam1p F0501 318 NP_015277.1 (Saccharomyces cerevisiae) DNA polymerase III subunit β (Cryptococcus F0701 433 XP_772271.1 neoformans) Protein tyrosine phosphatases (Cryptococcus F0704 411 XP_774214.1 neoformans) Serine/Threonine protein kinases (Aspergillus F0801 309 XP_751359.1 fumigatus) F1304 490 GAL4-like DNA-binding domain (Gibberella XP_386500.1 zeae) F1700 640 tRNA modification GTPase (Candida glabrata) XP_449676.1 tRNA modification GTPase (Candida F1902 637 XP_002418056.1 dubliniensis) Cell structure A0800 199 LysM (Aspergillus nidulans) XP_682658.1 A1001 709 Hydrophobin (Gibberella zeae) XP_384136.1 B0501 1054 O-Glycosyl hydrolases (Laccaria bicolor) XP_001878748.1 B1100 819 Glucan synthase (Ustilago maydis) XP_757786.1 Phosphatidylinositol N-acetylglucosaminyl B1206 409 XP_002562782.1 transferase (Penicillium chrysogenum) Haemolysin-III related (Lachancea B1401 254 XP_002552634.1 thermotolerans) D1600 711 α-tubulin (Cryptococcus neoformans) XP_568869.1 1, 3 β-glucan synthase component (Ustilago D1700 648 XP_757786.1 maydis) O-mannosyl transferase (Lachancea E0301 532 XP_002554648.1 thermotolerans) Eukaryotic membrane protein (Aspergillus E0402 390 XP_659482.1 nidulans) F0803 511 Dynamitin (Penicillium chrysogenum) XP_002564327.1 Unknown A0801 126 No significant similarity found - Hypothetical protein An14g01900 (Aspergillus A1500 375 XP_001400816.1 niger) B0703 265 No significant similarity found - B1002 322 Hypothetical protein UM03492.1 (Ustilago XP_759639.1 maydis) B1003 236 No significant similarity found - Hypothetical protein AN7386.2 (Aspergillus C1011 355 XP_680655.1 nidulans) E0500 761 Hypothetical protein (Encephalitozoon cuniculi) NP_597351.1 E0602 239 No significant similarity found - E1300 253 No significant similarity found - F0600 496 Hypothetical protein (Cryptococcus neoformans) XP_569554.1 F0702 524 Hypothetical protein (Phaeosphaeria nodorum) XP_001794814.1 F1301 422 No significant similarity found - Other genes B0101 454 Eukaryotic aspartyl protease (Laccaria bicolor) XP_001880663.1 B1402 359 LCCL domain (Aspergillus niger) XP_001391575.1 Phosphatidylinositol 4-phosphate 5-kinase D1112 508 XP_001383052.2 (Pichia stipitis) Proteasome alpha-type 6 (Cryptococcus D1302 448 XP_568975.1 neoformans) E2000 552 Ankyrin repeats (Vanderwaltozyma polyspora) XP_001646456.1 Disulfide-isomerase precursor (Cryptococcus F0203 696 XP_566486.1 neoformans) Transporter B0202 658 Sugar transporter (Aspergillus clavatus) XP_001272286.1 Cytosine/uracil/thiamine/allantoin permeases C1003 424 XP_445163.1 (Candida glabrata) ATP-binding cassette transporters (Pichia E0104 852 XP_001383771.1 stipitis) Transmembrane amino acid transporter protein F0202 571 XP_759838.1 (Ustilago maydis) a The percentages are based on BlastX searches of the Genbank database. The other numbers in this column show the number of amino acids (query/subject) that were compared