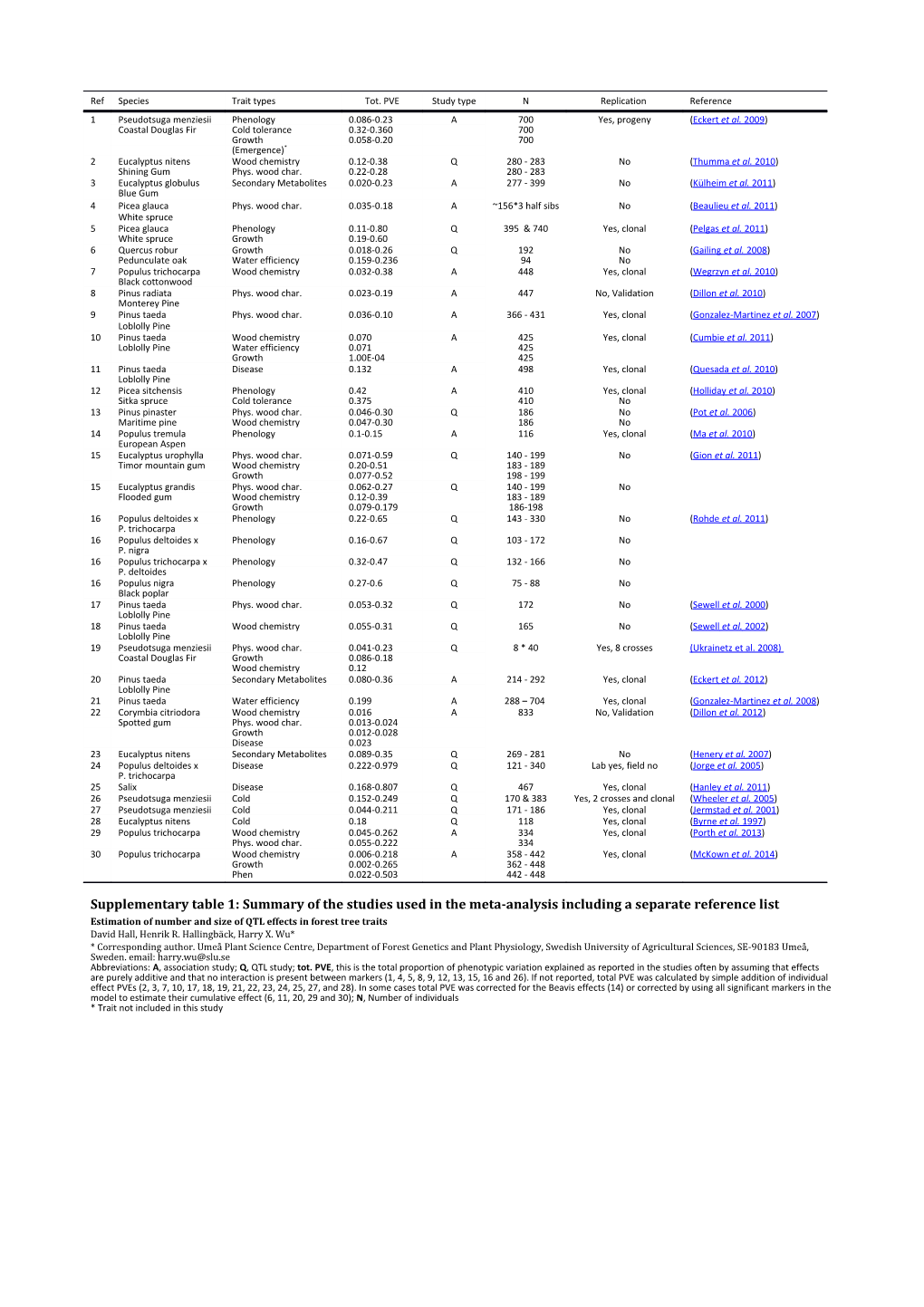
Ref Species Trait types Tot. PVE Study type N Replication Reference 1 Pseudotsuga menziesii Phenology 0.086-0.23 A 700 Yes, progeny ( Eckert et al. 2009) Coastal Douglas Fir Cold tolerance 0.32-0.360 700 Growth 0.058-0.20 700 (Emergence)* 2 Eucalyptus nitens Wood chemistry 0.12-0.38 Q 280 - 283 No ( Thumma et al. 2010) Shining Gum Phys. wood char. 0.22-0.28 280 - 283 3 Eucalyptus globulus Secondary Metabolites 0.020-0.23 A 277 - 399 No ( Külheim et al. 2011) Blue Gum 4 Picea glauca Phys. wood char. 0.035-0.18 A ~156*3 half sibs No ( Beaulieu et al. 2011) White spruce 5 Picea glauca Phenology 0.11-0.80 Q 395 & 740 Yes, clonal ( Pelgas et al. 2011) White spruce Growth 0.19-0.60 6 Quercus robur Growth 0.018-0.26 Q 192 No ( Gailing et al. 2008) Pedunculate oak Water efficiency 0.159-0.236 94 No 7 Populus trichocarpa Wood chemistry 0.032-0.38 A 448 Yes, clonal ( Wegrzyn et al. 2010) Black cottonwood 8 Pinus radiata Phys. wood char. 0.023-0.19 A 447 No, Validation ( Dillon et al. 2010) Monterey Pine 9 Pinus taeda Phys. wood char. 0.036-0.10 A 366 - 431 Yes, clonal ( Gonzalez-Martinez et al. 2007) Loblolly Pine 10 Pinus taeda Wood chemistry 0.070 A 425 Yes, clonal ( Cumbie et al. 2011) Loblolly Pine Water efficiency 0.071 425 Growth 1.00E-04 425 11 Pinus taeda Disease 0.132 A 498 Yes, clonal ( Quesada et al. 2010) Loblolly Pine 12 Picea sitchensis Phenology 0.42 A 410 Yes, clonal ( Holliday et al. 2010) Sitka spruce Cold tolerance 0.375 410 No 13 Pinus pinaster Phys. wood char. 0.046-0.30 Q 186 No ( Pot et al. 2006) Maritime pine Wood chemistry 0.047-0.30 186 No 14 Populus tremula Phenology 0.1-0.15 A 116 Yes, clonal ( Ma et al. 2010) European Aspen 15 Eucalyptus urophylla Phys. wood char. 0.071-0.59 Q 140 - 199 No ( Gion et al. 2011) Timor mountain gum Wood chemistry 0.20-0.51 183 - 189 Growth 0.077-0.52 198 - 199 15 Eucalyptus grandis Phys. wood char. 0.062-0.27 Q 140 - 199 No Flooded gum Wood chemistry 0.12-0.39 183 - 189 Growth 0.079-0.179 186-198 16 Populus deltoides x Phenology 0.22-0.65 Q 143 - 330 No ( Rohde et al. 2011) P. trichocarpa 16 Populus deltoides x Phenology 0.16-0.67 Q 103 - 172 No P. nigra 16 Populus trichocarpa x Phenology 0.32-0.47 Q 132 - 166 No P. deltoides 16 Populus nigra Phenology 0.27-0.6 Q 75 - 88 No Black poplar 17 Pinus taeda Phys. wood char. 0.053-0.32 Q 172 No ( Sewell et al. 2000) Loblolly Pine 18 Pinus taeda Wood chemistry 0.055-0.31 Q 165 No ( Sewell et al. 2002) Loblolly Pine 19 Pseudotsuga menziesii Phys. wood char. 0.041-0.23 Q 8 * 40 Yes, 8 crosses (Ukrainetz et al. 2008) Coastal Douglas Fir Growth 0.086-0.18 Wood chemistry 0.12 20 Pinus taeda Secondary Metabolites 0.080-0.36 A 214 - 292 Yes, clonal ( Eckert et al. 2012) Loblolly Pine 21 Pinus taeda Water efficiency 0.199 A 288 – 704 Yes, clonal ( Gonzalez-Martinez et al. 2008) 22 Corymbia citriodora Wood chemistry 0.016 A 833 No, Validation ( Dillon et al. 2012) Spotted gum Phys. wood char. 0.013-0.024 Growth 0.012-0.028 Disease 0.023 23 Eucalyptus nitens Secondary Metabolites 0.089-0.35 Q 269 - 281 No ( Henery et al. 2007) 24 Populus deltoides x Disease 0.222-0.979 Q 121 - 340 Lab yes, field no ( Jorge et al. 2005) P. trichocarpa 25 Salix Disease 0.168-0.807 Q 467 Yes, clonal ( Hanley et al. 2011) 26 Pseudotsuga menziesii Cold 0.152-0.249 Q 170 & 383 Yes, 2 crosses and clonal ( Wheeler et al. 2005) 27 Pseudotsuga menziesii Cold 0.044-0.211 Q 171 - 186 Yes, clonal ( Jermstad et al. 2001) 28 Eucalyptus nitens Cold 0.18 Q 118 Yes, clonal ( Byrne et al. 1997) 29 Populus trichocarpa Wood chemistry 0.045-0.262 A 334 Yes, clonal ( Porth et al. 2013) Phys. wood char. 0.055-0.222 334 30 Populus trichocarpa Wood chemistry 0.006-0.218 A 358 - 442 Yes, clonal ( McKown et al. 2014) Growth 0.002-0.265 362 - 448 Phen 0.022-0.503 442 - 448
Supplementary table 1: Summary of the studies used in the meta-analysis including a separate reference list Estimation of number and size of QTL effects in forest tree traits David Hall, Henrik R. Hallingbäck, Harry X. Wu* * Corresponding author. Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences, SE-90183 Umeå, Sweden. email: [email protected] Abbreviations: A, association study; Q, QTL study; tot. PVE, this is the total proportion of phenotypic variation explained as reported in the studies often by assuming that effects are purely additive and that no interaction is present between markers (1, 4, 5, 8, 9, 12, 13, 15, 16 and 26). If not reported, total PVE was calculated by simple addition of individual effect PVEs (2, 3, 7, 10, 17, 18, 19, 21, 22, 23, 24, 25, 27, and 28). In some cases total PVE was corrected for the Beavis effects (14) or corrected by using all significant markers in the model to estimate their cumulative effect (6, 11, 20, 29 and 30); N, Number of individuals * Trait not included in this study References Table S1 Beaulieu J, Doerksen T, Boyle B, et al. (2011) Association Genetics of Wood Physical Traits in the Conifer White Spruce and Relationships With Gene Expression. Genetics 188, 197-U329. Byrne M, Murrell JC, Owen JV, Williams ER, Moran GF (1997) Mapping of quantitative trait loci influencing frost tolerance in Eucalyptus nitens. Theoretical and Applied Genetics 95, 975-979. Cumbie WP, Eckert A, Wegrzyn J, et al. (2011) Association genetics of carbon isotope discrimination, height and foliar nitrogen in a natural population of Pinus taeda L. Heredity 107, 105-114. Dillon SK, Brawner JT, Meder R, Lee DJ, Southerton SG (2012) Association genetics in Corymbia citriodora subsp. variegata identifies single nucleotide polymorphisms affecting wood growth and cellulosic pulp yield. New Phytologist 195, 596-608. Dillon SK, Nolan M, Li W, et al. (2010) Allelic Variation in Cell Wall Candidate Genes Affecting Solid Wood Properties in Natural Populations and Land Races of Pinus radiata. Genetics 185, 1477-1487. Eckert AJ, Bower AD, Wegrzyn JL, et al. (2009) Association Genetics of Coastal Douglas Fir (Pseudotsuga menziesii var. menziesii, Pinaceae). I. Cold-Hardiness Related Traits. Genetics 182, 1289-1302. Eckert AJ, Wegrzyn JL, Cumbie WP, et al. (2012) Association genetics of the loblolly pine (Pinus taeda, Pinaceae) metabolome. New Phytologist 193, 890-902. Gailing O, Langenfeld-Heyser R, Polle A, Finkeldey R (2008) Quantitative trait loci affecting stomatal density and growth in a Quercus robur progeny: implications for the adaptation to changing environments. Global Change Biology 14, 1934-1946. Gion JM, Carouche A, Deweer S, et al. (2011) Comprehensive genetic dissection of wood properties in a widely-grown tropical tree: Eucalyptus. Bmc Genomics 12. Gonzalez-Martinez SC, Huber D, Ersoz E, Davis JM, Neale DB (2008) Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 101, 19-26. Gonzalez-Martinez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB (2007) Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175, 399-409. Hanley SJ, Pei MH, Powers SJ, et al. (2011) Genetic mapping of rust resistance loci in biomass willow. Tree Genetics & Genomes 7, 597-608. Henery ML, Moran GF, Wallis IR, Foley WJ (2007) Identification of quantitative trait loci influencing foliar concentrations of terpenes and formylated phloroglucinol compounds in Eucalyptus nitens. New Phytologist 176, 82-95. Holliday JA, Ritland K, Aitken SN (2010) Widespread, ecologically relevant genetic markers developed from association mapping of climate-related traits in Sitka spruce (Picea sitchensis). New Phytologist 188, 501-514. Jermstad KD, Bassoni DL, Wheeler NC, et al. (2001) Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas- fir. II. Spring and fall cold-hardiness. Theoretical and Applied Genetics 102, 1152-1158. Jorge V, Dowkiw A, Faivre-Rampant P, Bastien C (2005) Genetic architecture of qualitative and quantitative Melampsora larici- populina leaf rust resistance in hybrid poplar: genetic mapping and QTL detection. New Phytologist 167, 113-127. Külheim C, Yeoh SH, Wallis IR, et al. (2011) The molecular basis of quantitative variation in foliar secondary metabolites in Eucalyptus globulus. New Phytologist 191, 1041-1053. Ma X-F, Hall D, Onge KRS, Jansson S, Ingvarsson PK (2010) Genetic Differentiation, Clinal Variation and Phenotypic Associations With Growth Cessation Across the Populus tremula Photoperiodic Pathway. Genetics 186, 1033-1044. McKown AD, Klápště J, Guy RD, et al. (2014) Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytologist 203, 535-553. Pelgas B, Bousquet J, Meirmans PG, Ritland K, Isabel N (2011) QTL mapping in white spruce: gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. Bmc Genomics 12. Porth I, Klapšte J, Skyba O, et al. (2013) Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytologist 200, 710-726. Pot D, Rodrigues JC, Rozenberg P, et al. (2006) QTLs and candidate genes for wood properties in maritime pine (Pinus pinaster Ait.). Tree Genetics & Genomes 2, 10-24. Quesada T, Gopal V, Cumbie WP, et al. (2010) Association Mapping of Quantitative Disease Resistance in a Natural Population of Loblolly Pine (Pinus taeda L.). Genetics 186, 677-686. Rohde A, Storme V, Jorge V, et al. (2011) Bud set in poplar - genetic dissection of a complex trait in natural and hybrid populations. New Phytologist 189, 106-121. Sewell MM, Bassoni DL, Megraw RA, Wheeler NC, Neale DB (2000) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). I. Physical wood properties. Theoretical and Applied Genetics 101, 1273-1281. Sewell MM, Davis MF, Tuskan GA, et al. (2002) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties. Theoretical and Applied Genetics 104, 214-222. Thumma BR, Southerton SG, Bell JC, et al. (2010) Quantitative trait locus (QTL) analysis of wood quality traits in Eucalyptus nitens. Tree Genetics & Genomes 6, 305-317. Wegrzyn JL, Eckert AJ, Choi M, et al. (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytologist 188, 515-532. Wheeler NC, Jermstad KD, Krutovsky K, et al. (2005) Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas- fir. IV. Cold-hardiness QTL verification and candidate gene mapping. Molecular Breeding 15, 145-156.