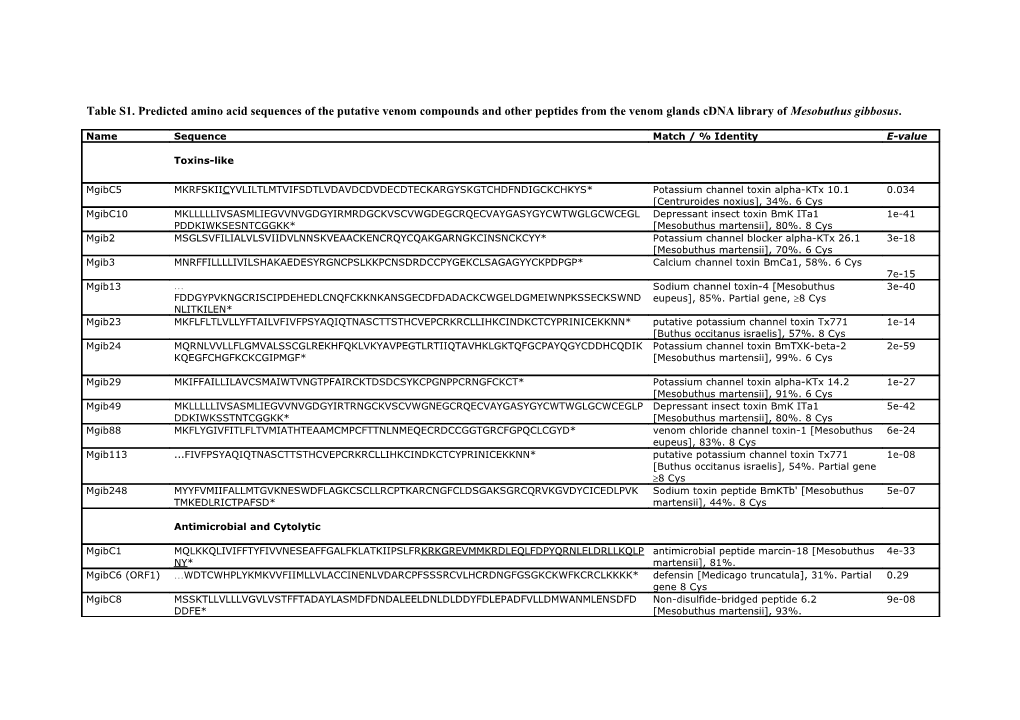
Table S1. Predicted amino acid sequences of the putative venom compounds and other peptides from the venom glands cDNA library of Mesobuthus gibbosus.
Name Sequence Match / % Identity E-value
Toxins-like
MgibC5 MKRFSKIICYVLILTLMTVIFSDTLVDAVDCDVDECDTECKARGYSKGTCHDFNDIGCKCHKYS* Potassium channel toxin alpha-KTx 10.1 0.034 [Centruroides noxius], 34%. 6 Cys MgibC10 MKLLLLLIVSASMLIEGVVNVGDGYIRMRDGCKVSCVWGDEGCRQECVAYGASYGYCWTWGLGCWCEGL Depressant insect toxin BmK ITa1 1e-41 PDDKIWKSESNTCGGKK* [Mesobuthus martensii], 80%. 8 Cys Mgib2 MSGLSVFILIALVLSVIIDVLNNSKVEAACKENCRQYCQAKGARNGKCINSNCKCYY* Potassium channel blocker alpha-KTx 26.1 3e-18 [Mesobuthus martensii], 70%. 6 Cys Mgib3 MNRFFILLLLIVILSHAKAEDESYRGNCPSLKKPCNSDRDCCPYGEKCLSAGAGYYCKPDPGP* Calcium channel toxin BmCa1, 58%. 6 Cys 7e-15 Mgib13 … Sodium channel toxin-4 [Mesobuthus 3e-40 FDDGYPVKNGCRISCIPDEHEDLCNQFCKKNKANSGECDFDADACKCWGELDGMEIWNPKSSECKSWND eupeus], 85%. Partial gene, 8 Cys NLITKILEN* Mgib23 MKFLFLTLVLLYFTAILVFIVFPSYAQIQTNASCTTSTHCVEPCRKRCLLIHKCINDKCTCYPRINICEKKNN* putative potassium channel toxin Tx771 1e-14 [Buthus occitanus israelis], 57%. 8 Cys Mgib24 MQRNLVVLLFLGMVALSSCGLREKHFQKLVKYAVPEGTLRTIIQTAVHKLGKTQFGCPAYQGYCDDHCQDIK Potassium channel toxin BmTXK-beta-2 2e-59 KQEGFCHGFKCKCGIPMGF* [Mesobuthus martensii], 99%. 6 Cys
Mgib29 MKIFFAILLILAVCSMAIWTVNGTPFAIRCKTDSDCSYKCPGNPPCRNGFCKCT* Potassium channel toxin alpha-KTx 14.2 1e-27 [Mesobuthus martensii], 91%. 6 Cys Mgib49 MKLLLLLIVSASMLIEGVVNVGDGYIRTRNGCKVSCVWGNEGCRQECVAYGASYGYCWTWGLGCWCEGLP Depressant insect toxin BmK ITa1 5e-42 DDKIWKSSTNTCGGKK* [Mesobuthus martensii], 80%. 8 Cys Mgib88 MKFLYGIVFITLFLTVMIATHTEAAMCMPCFTTNLNMEQECRDCCGGTGRCFGPQCLCGYD* venom chloride channel toxin-1 [Mesobuthus 6e-24 eupeus], 83%. 8 Cys Mgib113 ...FIVFPSYAQIQTNASCTTSTHCVEPCRKRCLLIHKCINDKCTCYPRINICEKKNN* putative potassium channel toxin Tx771 1e-08 [Buthus occitanus israelis], 54%. Partial gene 8 Cys Mgib248 MYYFVMIIFALLMTGVKNESWDFLAGKCSCLLRCPTKARCNGFCLDSGAKSGRCQRVKGVDYCICEDLPVK Sodium toxin peptide BmKTb' [Mesobuthus 5e-07 TMKEDLRICTPAFSD* martensii], 44%. 8 Cys
Antimicrobial and Cytolytic
MgibC1 MQLKKQLIVIFFTYFIVVNESEAFFGALFKLATKIIPSLFRKRKGREVMMKRDLEQLFDPYQRNLELDRLLKQLP antimicrobial peptide marcin-18 [Mesobuthus 4e-33 NY* martensii], 81%. MgibC6 (ORF1) …WDTCWHPLYKMKVVFIIMLLVLACCINENLVDARCPFSSSRCVLHCRDNGFGSGKCKWFKCRCLKKKK* defensin [Medicago truncatula], 31%. Partial 0.29 gene 8 Cys MgibC8 MSSKTLLVLLLVGVLVSTFFTADAYLASMDFDNDALEELDNLDLDDYFDLEPADFVLLDMWANMLENSDFD Non-disulfide-bridged peptide 6.2 9e-08 DDFE* [Mesobuthus martensii], 93%. MgibC9 MNKKTLLVIFFVTMLIIDEVNSIRWGSLFKRVWKSKLARKLRSKGKSLLKDYANRVLSGGPEEEAAPPAERKR Bradykinin-potentiating peptide NDBP6 2e-13 * [Lychas mucronatus], 85%. Mgib253 MSSKTLLVLLLVGVLVSTFFTADAYPASMDFDNDALEELDNLDLDDYFDLEPADFVLLDMWANMLENSDFD Non-disulfide-bridged peptide 6.2 1e-26 DDFEY* [Mesobuthus martensii], 94%.
Other venom components
MgibC11 MTLTNGLSLRTIFITLLLLLPPHLLATTASITKIQTRNRTRKVRGFILLSEAAENRD* venom protein Txlp2 [Hottentotta judaicus], 1e-12 79%. Mgib223 MKRFLVFSILFQTVFCMKTSDQVGIITYQGVPRRERKCILGPWIHLEDGSVFHDSDRCEIKTCHITAQKAYLE venom peptide [Hottentotta judaicus], 28%. VQSCRYKVNCERQILEPYFPHCCPTSPKCT* 8 cys 5.8 Mgib277 ...WCGAGSSAKHEDELGRFNDTDSCCRHHDHCHDNIKGKETKYGLKNKDSTTMSHCDCDEEFYACLKTVD phospholipase A2D precursor [Tribolium 1e-37 SIISNNVGNMFFNFLQKKCFREDYPIKRCVKKSLIRRRCKKYELDRTKPKIWQIFDPKEY* castaneum], 49%. Partial gene. 10 Cys
Cellular Proteins
MgibC6 (ORF2) MCELDILHDSLYQFCPELHLKRLNSLTLACHALLDCKTLTLTELGRNLPTKARTKHNIKRIDRLLGNRHLHKER transposase of Tn10 [Shigella flexneri 2b], LAVYRWHASFICSGNTMPIVLVDWSDIREQKRLMVLRASVALHGRSVTLYEKAFPLSEQCSKKAHDQFLADL 100% ASILPSNTTPLIVSDAGFKVPWYKSVEKLGWYWLSRVRGKVQYADLGAENWKPISNLHDMSSSHSKTLGYK RLTKSNPISCQILLYKSRSKGRKNQRSTRTHCHHPSPKIYSASAKEPWILATNLPVEIRTPKQLVNIYSKRMQI EETFRDLKSPAYGLGLRHSRTSSSERFDIMLLIALMLQLTCWLAGVHAQKQGWDKHFQANTVRNRNVLSTV RLGMEVLRHSGYTITREDSLVAATLLTQNLFTHGYVLGKL* MgibC7 MKLILLLIVMGILAISKCSVGNTLCELSRNQKIVLLECLESYLTEEQKASEYQFYQCLGYDSVIDYYEEICGLSE ribonuclease R [Coxiella burnetii RSA 331], 0.61 KEQEELHLSYIKCHNEMTPYSGSATDEDARNCLNNAIESEQ* 33%. Partial gene Mgib18 MATTSKPEDHRKKWDRDEFERLAHERILEEYELEKKAKDKQPPIRREYLKPRDYRVDLESKLGKTTVITKTTP zinc finger matrin-type protein 2-like [Oryzias 3e-89 ASQAGGYYCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLEQVKKRFEMNRKKLDEKKKEYDFE latipes] Actinopterygii, 69%. QRMQELRDEEEKLKEYRRERRKERKRKCQDSRDDDIDPEMAAIMGFSEFGSSKK* Mgib26 … NADH dehydrogenase subunit 3 [Mesobuthus 3e-43 VLLLVLVFRILLRSIILFVGGVISKKLEVEKEKGSVYECGFESRKRARVPFSLQFFMVGVIFLIFDVEIVLIMPVP gibbosus], 90%. LEAWLRGGEVLLFFFFVFLLLFGLFFE* Mgib36 …MLISKIIKRIDWFPSVFKYFYRSERGYRKGLRRKPISVISENKNEDAKQIFKRDIMENLKQE... Monogalactosyldiacylglycerol synthase, partial 4.1 [Megasphaera sp. NM10], 43% Mgib104 GGILRNLHLNGARIFFVCLYFHIGRGIYFGSFKFYFTwITGVVIFLLTIITAFLGYVLPwGQMSFwGATVITNLV cytochrome b [Mesobuthus gibbosus], partial, SAVPYIGSEVVQWLwGGFSVDNPTLTRFFSFHFLLPFVILGRVIIHLIFLHERGSRNPLGVKRDFDKIPFHPYF 95% SLKDLLGALFLIIFLIFISLLFPNFLSDPENCIPANPLVTPVHIQPEwYFLFAYAILRSIPNKLGGVIALIMSVLILIL LSFSTSKSRAFSFRIGSRSLFwILVNIFILLTwIGARPVEFPYIMLGQGLS Mgib142 MGIVTVHKKKSNLKMNKGTMFLSFILILLIVDGIKSYNIEREEELEERLLDDDLKEELEENRNELRQKRDRYLP Adhesive plaque matrix protein, partial [Bos 2e-17 TVQKRERYLPTVQKRERYLPTVQKRERYLPTVQKRERYLPTVQKRERYLPTVQKRERYLPTVQKRERYLPTVQ grunniens mutus], 20% KRERYLPTR* Mgib263 EST MPPKQDQKKKDTKGTTKKKEGASSGGKAKKKKWSKGKVRDKLNNLVLFDKATYEKLLKEVPSYKLITPSVV putative 40S ribosomal protein S25 6e-51 SERLKVRGSLARRALEELRQKGLIKQVVKHHSQIIYTRTTKADDST* [Dolomedes mizhoanus], 87% Mgib264 MSWQAYVDNQICAQVSCRLAVIAGLQDGAIWAKFEKDMPKPVTQQELKLIADTMRTNPNSFTESGIRLGED Blo t profilin allergen [Latrodectus hesperus], 2e-76 KYYCLHAENSLLRGRKGSSALIVVATNTCLLVAATTDGFPPGQLNAVVEKLGDYLRSNNY* 84%
Unkown (hypothetical)
MgibC3 GVCSTFKIFHDLSSNRCKPGWFLS hypothetical protein 11, partial [Urodacus 7e-05 yaschenkoi], 92%. MgibC4 GVCSTFKIFHDLSSNRCKPGWFLS hypothetical protein 11, partial [Urodacus 6e-05 yaschenkoi], 92%. Mgib1 MKNLAIITLSLISLSTSFSLFPSLSLPSVPLYPSSLIPLFMKDPVKEITKIYCVLKNVNIARSCARENNQGKISDF hypothetical secreted protein [Hottentotta 4e-32 FRKCMQKITSLKTLDEMRDFYCNKMNLEETFKAKACLDPGIMNMVLTDATFVPTILNCLFKAKNSEK* judaicus], 45%. 6 Cys Mgib45 MKPVQSVVLFLLLLALHASLSARIARQAEDLDDENNEVPQDEDSALDPDEDSEEDDDDGFSFRFSWNPFQN hypothetical secreted protein [Hottentotta 3e-42 FPDIFKQMRDNMNWIYSNLFNETSNLPEVYKNVSSEIVTIGGSKFNVSKTIIKKADNNSQVVISSVSMDQVN judaicus], 77%. KKKK* Mgib72 EST …SSSLPEAVLSVFRLLPLSSISLTLRFTVILRTSGITSHKTCYPDVPKMPGTIKKTSLLALCP* hypothetical protein [Plasmodium berghei 0.070 strain ANKA], 46%. Partial gene Mgib95 MMESLSCVGLSYALIEYIVNSDLSLFYPQLKSCLFLASCEECILDVESYCSFSPPNKSVAVKEHVLAACKIYVE conserved hypothetical protein [Ixodes 6e-69 GREGVIILDPGYHVGIPIVAMKDGLYPHKGWFVQSETPKSKKEYCYQLKGDYVHWTVRETRNGKEETWCNL scapularis], 42%. IYIKKKFLSYISVSEKRNLVFNFRTLVARDEKQPVGGFYCNLEGDEKFTLFFKDYLGKRIEIKIPFGYFKGSRNN NEYESAIRKCAVQIKTNSALLVGMMTQLVEAYYDKEFMLPVNEINREIDED* Mgib99 EST … hypothetical protein CAPTEDRAFT_188127 2e-05 EEICHMLTKYIKLRKLPELFSKLFASLENNSTEKNLIYPAEFLQDLSSQISYLPAGQILEIWKICLVNGSKLLNDI [Capitella teleta] Polychaeta, 25% . Partial KKSKNEISIKCLNMIAEILRIFILHFKIDRDICLSSKKQTELENITVFIKKFMNICIKKNIQADLGYSILSLCYVW gene GELHLSMKHYNRFYQFKIEFTKEDKGLFDLNAIHYYFTSQEWTDLLNNFENIKENKIQY… Mgib222 MKISIALVLGMIACSFVCINGGDFDQYCSIPSNIRELVWNCIYIYLPQSEKTVFSQMSQCFGVSTFDKVINQF hypothetical protein [Pandinus cavimanus], 5e-05 CGLSDDEQETVALQHEACLSEIDFDNFHGPTVSQYQTQSCIQRRLG* 27%. Mgib267 EST …VPFFFSGIFFFEHAQFLHYQVFKKMFHSHISIMNIFW* hypothetical protein [Vibrio splendidus]. 0.53 Partial gene
No Match
Mgib55 EST MVISYLYFLFEIFLEGSVKRGGGVFFGGVFENYVMGGREPLFAGKVALCCFKKNFFSVF* Mgib16 EST MGGVSPLLAPPVFPFGVPVFFFGGIWGGKNFPPGNPFAPVFPPVPPPRGGSWGSPPGGPTRGPGGGPSFPR GAKFCGSLRTPPKNFF*