1
Supplementary Data
Detection of a point mutation.
The structures of the transition molecules and the model molecular markers used for detection of point mutation (Supplementary Fig. S2) are given in Table S8. In the experiment, each transition molecule was taken at 1 M and total concentration of the model markers was set to 2 M. The ratio between the sequences was gradually varied as shown in Supplementary Fig. S3b. The mixture of the transition molecules and the model markers was equilibrated for 10 min at 15 oC; one-step computation was initiated by addition of 1 M of FokI enzyme and proceeded at 15 oC for 30 min prior to quenching and analysis by denaturing PAGE.
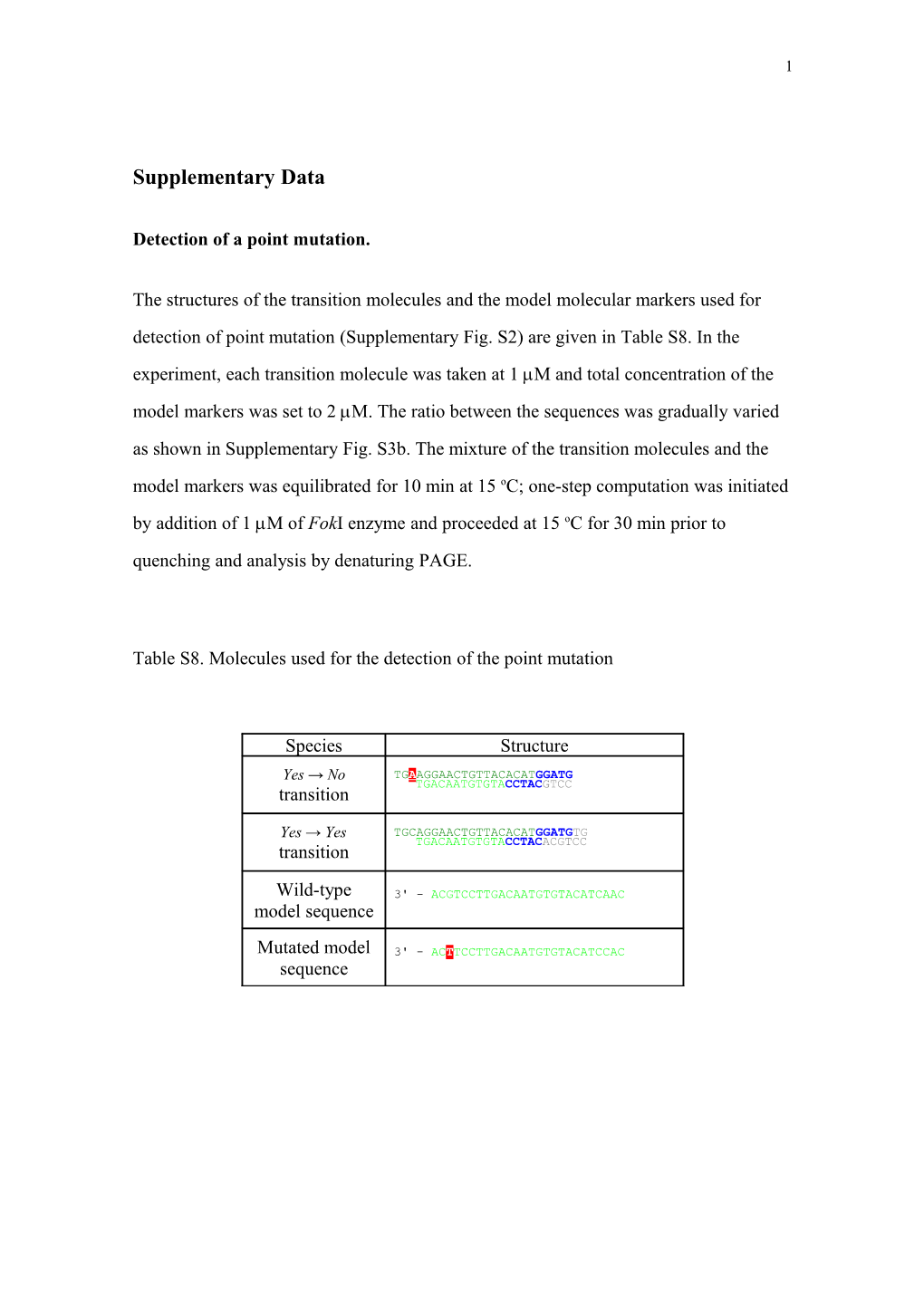
Table S8. Molecules used for the detection of the point mutation
Species Structure
Yes → No TGAAGGAACTGTTACACATGGATG TGACAATGTGTACCTACGTCC transition
Yes → Yes TGCAGGAACTGTTACACATGGATGTG TGACAATGTGTACCTACACGTCC transition
Wild-type 3' - ACGTCCTTGACAATGTGTACATCAAC model sequence
Mutated model 3' - ACTTCCTTGACAATGTGTACATCCAC sequence 2
Adjusting confidence in a positive diagnosis for various concentrations of the molecular indicator.
The experiment described in Supplementary Figs. S3 (c-e) was performed using the
ASCL1 SCLC diagnostic molecule. The computation advanced by Yes Yes and Yes
GRIA2 Yes transitions, then branched on INSM1↑ symbol due to regulation by the
PTTG1 INSM ssDNA model disease marker and proceeded to completion via Yes Yes
PTTG1 and No No transitions, to reflect the Yes/No ratio obtained at the branching point. All transition molecules except the regulated pair and the diagnostic string were taken at 1 M concentration, and FokI enzyme was taken at 5.4 M concentration. To improve the regulation pattern, a single-stranded protecting oligonucleotide for the Yes
INSM1 Yes transition molecule was taken at the same concentration (indicated in the
Supplementary Fig. S3) as the active regulated transitions.
Release of ssDNA molecule with a sequence identical to the approved antisense drug
In order to introduce the N-mer ssDNA drug into a diagnostic molecule, we form an (N- 4)-mer loop due to the restriction pattern of FokI. To design a drug suppressor we use a complementary ssDNA sequence for the ssDNA drug loops that are equal to or less than 12 nucleotides (N ≤ 16). For longer ssDNA drug loops, this sequence is furnished with additional nucleotides at its 3'-terminus that are complementary to 5'-region of the original single stranded loop and thus decrease the size of the loop to 12 nt or less. This design prevents an active drug from hybridizing to the inactive drug suppressor and vice versa.
We demonstrate this principle by designing a drug-releasing moiety that administers a ssDNA molecules that has the same nucleotide sequence as the Vitravene® (FDA 3
approved antisense drug) which is 21 nucleotides long. This drug requires a 17-mer loop, therefore the drug suppressor-release moiety was designed to have a 8-mer loop by elongating the drug suppressor to be 28 nt (instead of 21 nt) long. As the drug molecule did not contain an internal label, we used a labelled drug suppressor as a probe to indicate drug release.
Supplementary Fig. S5 shows a formation of the suppressed drug upon positive diagnosis indicating release of an active drug, and the absence of this species upon negative diagnosis meaning the contrary. A small amount of undesired cross- hybridization product between the drug suppressor probe and the unprocessed diagnostic molecule suggests that the design should be improved further to eliminate these interactions completely.
To perform the experiment, the diagnostic molecule, containing one symbolic indicator GSTP1↓, was incubated at a final concentration of 1 M in NEB4 buffer containing 1 M synthetic (28-mer) 32P-labelled drug suppressor. Positive or negative transition for GSTP1↓ (1 M) and Yes-verification-transitions (2 M) were added. The computation was initiated by addition of 5.4 M of FokI to a 10 l total volume. After incubating the mixture for 30 min at 15oC the reactions were quenched with 10 mM EDTA, mixed with the loading buffer and analyzed by native PAGE (20%). The exact structures of the molecular species used in the experiments are given in supplementary table S9. 4
Table S9. Molecules used in Vitravene® release experiment.
Species Structure
Drug suppressor CGCAAGAAGAAGAGCAAACGCTTCTTCT probe TTGCTCT GGTGCGCGACGCTCGACGCTCGACGCTCGCGT T Diagnostic molecule GCGCTGCGAGCTGCGAGCTGCGAGCGCA C GCGTTCTT