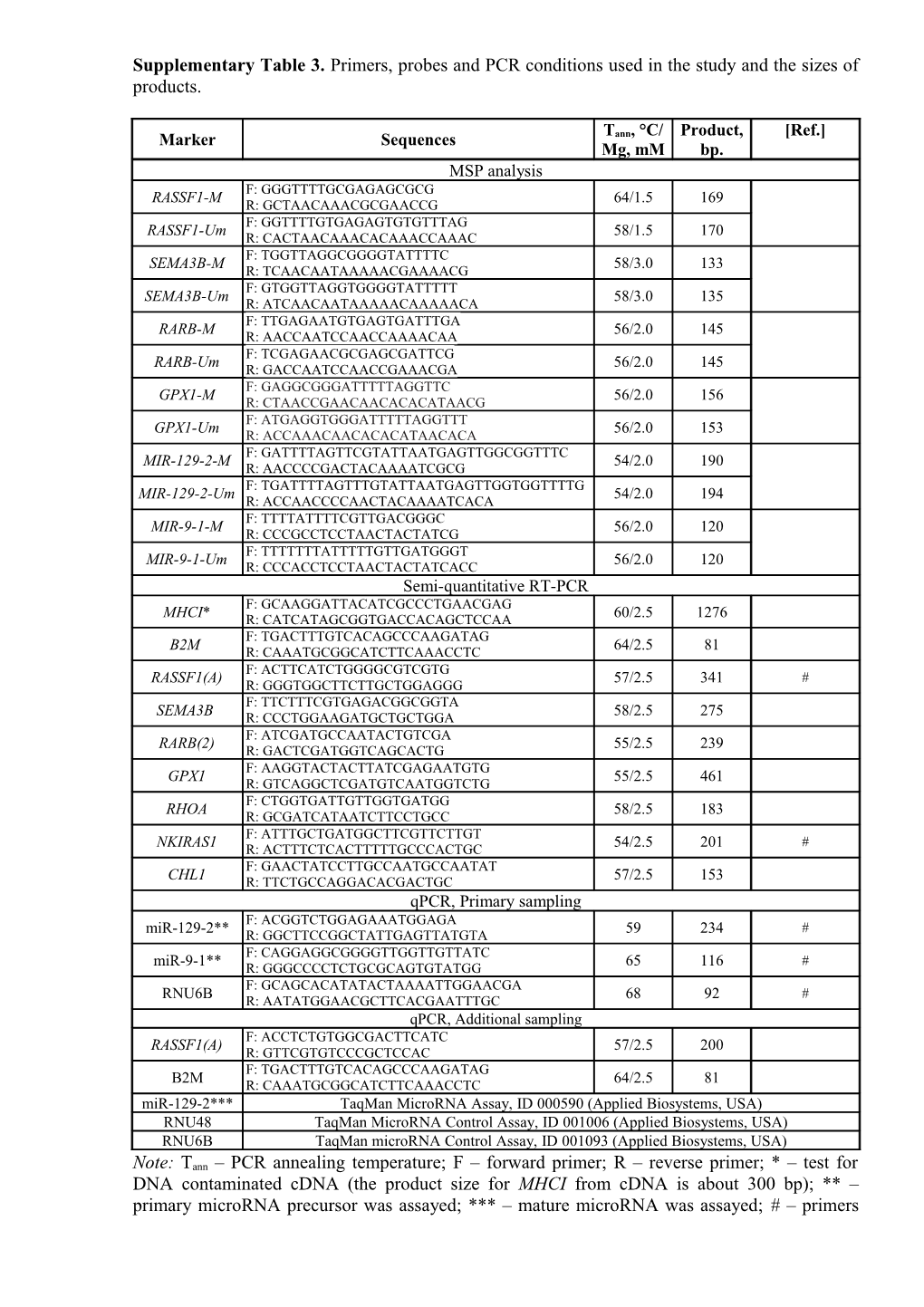
Supplementary Table 3. Primers, probes and PCR conditions used in the study and the sizes of products.
T , °C/ Product, [Ref.] Marker Sequences ann Mg, mM bp. MSP analysis F: GGGTTTTGCGAGAGCGCG RASSF1-M 64/1.5 169 R: GCTAACAAACGCGAACCG F: GGTTTTGTGAGAGTGTGTTTAG RASSF1-Um 58/1.5 170 R: CACTAACAAACACAAACCAAAC F: TGGTTAGGCGGGGTATTTTC SEMA3B-M 58/3.0 133 R: TCAACAATAAAAACGAAAACG F: GTGGTTAGGTGGGGTATTTTT SEMA3B-Um 58/3.0 135 R: ATCAACAATAAAAACAAAAACA F: TTGAGAATGTGAGTGATTTGA RARB-M 56/2.0 145 R: AACCAATCCAACCAAAACAA F: TCGAGAACGCGAGCGATTCG RARB-Um 56/2.0 145 R: GACCAATCCAACCGAAACGA F: GAGGCGGGATTTTTAGGTTC GPX1-M 56/2.0 156 R: CTAACCGAACAACACACATAACG F: ATGAGGTGGGATTTTTAGGTTT GPX1-Um 56/2.0 153 R: ACCAAACAACACACATAACACA F: GATTTTAGTTCGTATTAATGAGTTGGCGGTTTC MIR-129-2-M 54/2.0 190 R: AACCCCGACTACAAAATCGCG F: TGATTTTAGTTTGTATTAATGAGTTGGTGGTTTTG MIR-129-2-Um 54/2.0 194 R: ACCAACCCCAACTACAAAATCACA F: TTTTATTTTCGTTGACGGGC MIR-9-1-M 56/2.0 120 R: CCCGCCTCCTAACTACTATCG F: TTTTTTTATTTTTGTTGATGGGT MIR-9-1-Um 56/2.0 120 R: CCCACCTCCTAACTACTATCACC Semi-quantitative RT-PCR F: GCAAGGATTACATCGCCCTGAACGAG MHCI* R: CATCATAGCGGTGACCACAGCTCCAA 60/2.5 1276 F: TGACTTTGTCACAGCCCAAGATAG B2M R: CAAATGCGGCATCTTCAAACCTC 64/2.5 81 F: ACTTCATCTGGGGCGTCGTG RASSF1(A) 57/2.5 341 # R: GGGTGGCTTCTTGCTGGAGGG F: TTCTTTCGTGAGACGGCGGTA SEMA3B R: CCCTGGAAGATGCTGCTGGA 58/2.5 275 F: ATCGATGCCAATACTGTCGA RARB(2) R: GACTCGATGGTCAGCACTG 55/2.5 239 F: AAGGTACTACTTATCGAGAATGTG GPX1 55/2.5 461 R: GTCAGGCTCGATGTCAATGGTCTG F: CTGGTGATTGTTGGTGATGG RHOA 58/2.5 183 R: GCGATCATAATCTTCCTGCC F: ATTTGCTGATGGCTTCGTTCTTGT # NKIRAS1 R: ACTTTCTCACTTTTTGCCCACTGC 54/2.5 201 F: GAACTATCCTTGCCAATGCCAATAT CHL1 R: TTCTGCCAGGACACGACTGC 57/2.5 153 qPCR, Primary sampling F: ACGGTCTGGAGAAATGGAGA # miR-129-2** R: GGCTTCCGGCTATTGAGTTATGTA 59 234 F: CAGGAGGCGGGGTTGGTTGTTATC # miR-9-1** R: GGGCCCCTCTGCGCAGTGTATGG 65 116 F: GCAGCACATATACTAAAATTGGAACGA # RNU6B R: AATATGGAACGCTTCACGAATTTGC 68 92 qPCR, Additional sampling F: ACCTCTGTGGCGACTTCATC RASSF1(A) 57/2.5 200 R: GTTCGTGTCCCGCTCCAC F: TGACTTTGTCACAGCCCAAGATAG B2M R: CAAATGCGGCATCTTCAAACCTC 64/2.5 81 miR-129-2*** TaqMan MicroRNA Assay, ID 000590 (Applied Biosystems, USA) RNU48 TaqMan MicroRNA Control Assay, ID 001006 (Applied Biosystems, USA) RNU6B TaqMan microRNA Control Assay, ID 001093 (Applied Biosystems, USA)
Note: Tann – PCR annealing temperature; F – forward primer; R – reverse primer; * – test for DNA contaminated cDNA (the product size for MHCI from cDNA is about 300 bp); ** – primary microRNA precursor was assayed; *** – mature microRNA was assayed; # – primers were chosen using PrimerSelect from a software package Lasergene (http://www.dnastar.com/t- primerselect.aspx).
References Bandres, E., Agirre, X., Bitarte, N., Ramirez, N., Zarate, R., Roman-Gomez, J., Prosper, F. and Garcia-Foncillas, J., 2009. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 125, 2737-43. doi: 10.1002/ijc.24638. Beckedorff, F.C., Ayupe, A.C., Crocci-Souza, R., Amaral, M.S., Nakaya, H.I., Soltys, D.T., Menck, C.F., Reis, E.M. and Verjovski-Almeida, S., 2013. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet 9, e1003705. doi: 10.1371/journal.pgen.1003705. Burbee, D.G., Forgacs, E., Zochbauer-Muller, S., Shivakumar, L., Fong, K., Gao, B., Randle, D., Kondo, M., Virmani, A., Bader, S., Sekido, Y., Latif, F., Milchgrub, S., Toyooka, S., Gazdar, A.F., Lerman, M.I., Zabarovsky, E., White, M. and Minna, J.D., 2001. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93, 691-9. doi. Horiuchi, A., Imai, T., Wang, C., Ohira, S., Feng, Y., Nikaido, T. and Konishi, I., 2003. Up- regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab Invest 83, 861-70. doi. Li, S., Yan, T., Yang, J.Q., Oberley, T.D. and Oberley, L.W., 2000. The role of cellular glutathione peroxidase redox regulation in the suppression of tumor cell growth by manganese superoxide dismutase. Cancer Res 60, 3927-39. doi. Loginov, V.I., Dmitriev, A.A., Senchenko, V.N., Pronina, I.V., Khodyrev, D.S., Kudryavtseva, A.V., Krasnov, G.S., Gerashchenko, G.V., Chashchina, L.I., Kazubskaya, T.P., Kondratieva, T.T., Lerman, M.I., Angeloni, D., Braga, E.A. and Kashuba, V.I., 2015. Tumor Suppressor Function of the SEMA3B Gene in Human Lung and Renal Cancers. PLoS One 10, e0123369. doi: 10.1371/journal.pone.0123369. Lujambio, A., Calin, G.A., Villanueva, A., Ropero, S., Sanchez-Cespedes, M., Blanco, D., Montuenga, L.M., Rossi, S., Nicoloso, M.S., Faller, W.J., Gallagher, W.M., Eccles, S.A., Croce, C.M. and Esteller, M., 2008. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 105, 13556-61. doi: 10.1073/pnas.0803055105. Min, S.Y., Kim, H.S., Jung, E.J., Jung, E.J., Jee, C.D. and Kim, W.H., 2012. Prognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res 32, 3169-75. doi. Senchenko, V.N., Krasnov, G.S., Dmitriev, A.A., Kudryavtseva, A.V., Anedchenko, E.A., Braga, E.A., Pronina, I.V., Kondratieva, T.T., Ivanov, S.V., Zabarovsky, E.R. and Lerman, M.I., 2011. Differential expression of CHL1 gene during development of major human cancers. PLoS One 6, e15612. doi: 10.1371/journal.pone.0015612. Widschwendter, M., Berger, J., Hermann, M., Muller, H.M., Amberger, A., Zeschnigk, M., Widschwendter, A., Abendstein, B., Zeimet, A.G., Daxenbichler, G. and Marth, C., 2000. Methylation and silencing of the retinoic acid receptor-beta2 gene in breast cancer. J Natl Cancer Inst 92, 826-32. doi. Yang, Q., Yoshimura, G., Nakamura, M., Nakamura, Y., Shan, L., Suzuma, T., Tamaki, T., Umemura, T., Mori, I. and Kakudo, K., 2001. Allelic loss of chromosome 3p24 correlates with tumor progression rather than with retinoic acid receptor beta2 expression in breast carcinoma. Breast Cancer Res Treat 70, 39-45. doi.