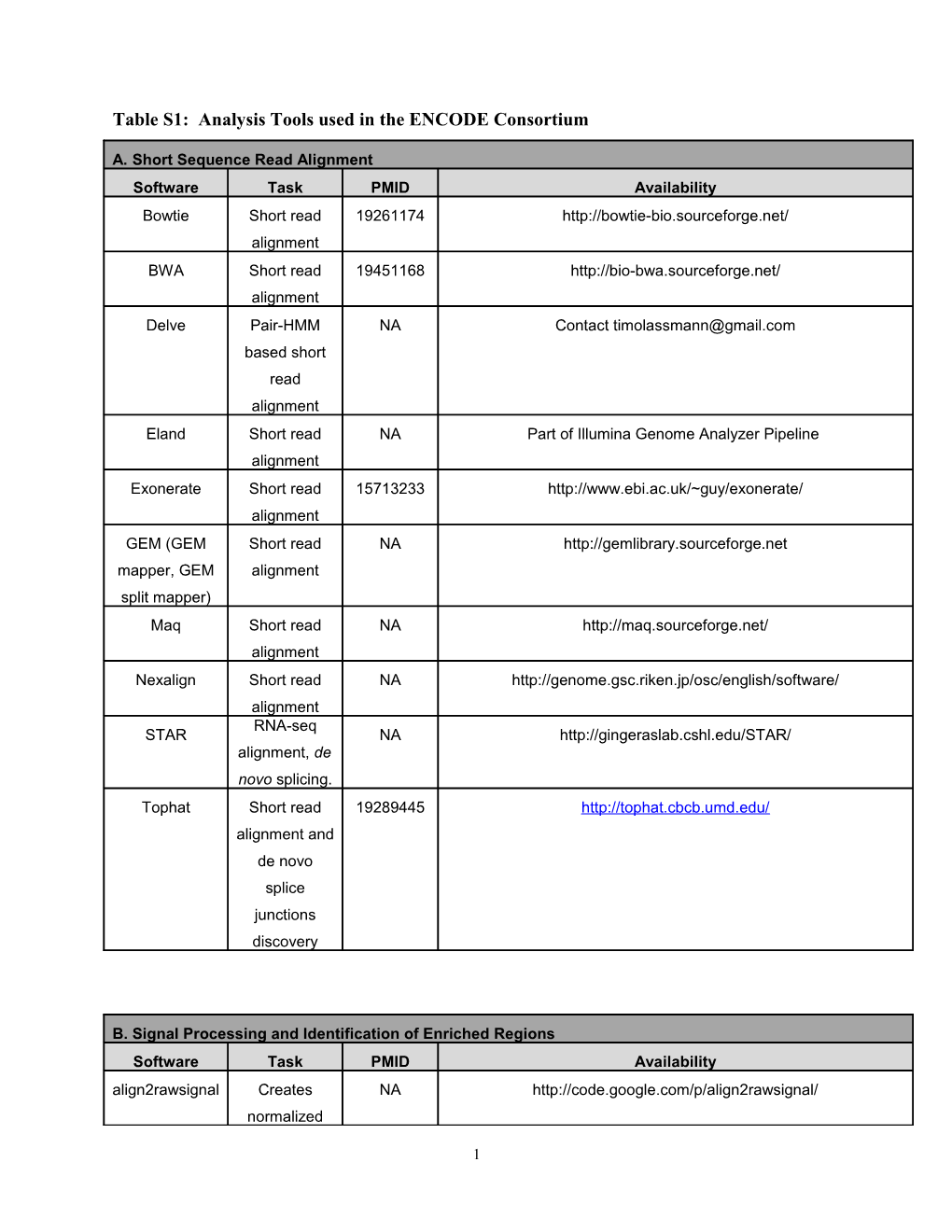
Table S1: Analysis Tools used in the ENCODE Consortium
A. Short Sequence Read Alignment Software Task PMID Availability Bowtie Short read 19261174 http://bowtie-bio.sourceforge.net/ alignment BWA Short read 19451168 http://bio-bwa.sourceforge.net/ alignment Delve Pair-HMM NA Contact [email protected] based short read alignment Eland Short read NA Part of Illumina Genome Analyzer Pipeline alignment Exonerate Short read 15713233 http://www.ebi.ac.uk/~guy/exonerate/ alignment GEM (GEM Short read NA http://gemlibrary.sourceforge.net mapper, GEM alignment split mapper) Maq Short read NA http://maq.sourceforge.net/ alignment Nexalign Short read NA http://genome.gsc.riken.jp/osc/english/software/ alignment RNA-seq STAR NA http://gingeraslab.cshl.edu/STAR/ alignment, de novo splicing. Tophat Short read 19289445 http://tophat.cbcb.umd.edu/ alignment and de novo splice junctions discovery
B. Signal Processing and Identification of Enriched Regions Software Task PMID Availability align2rawsignal Creates NA http://code.google.com/p/align2rawsignal/ normalized
1 signal from alignment Cufflinks Transcript 20436464 http://cufflinks.cbcb.umd.edu/manual.html reconstruction and quantification ERANGE Peak calling 18516045 http://woldlab.caltech.edu/rnaseq/ for ChIP-seq and RNA-seq Fluxcapacitor Transcript NA http://flux.sammeth.net/ quantification from RNAseq Fseq Peak calling 18784119 http://www.genome.duke.edu/labs/furey/software/fseq/ for DNase- seq, FAIRE- seq, and ChIP-seq Hotspot Peak calling NA http://www.uwencode.org/proj/hotspot-ptih for DNase- seq IQSeq Transcript NA http://archive.gersteinlab.org/proj/rnaseq/IQSeq quantification from RNA- seq data IDR Establishing NA http://www.encodestatistics.org/svn/idr/ reproducible replicate thresholds MACS Peak calling 18798982 http://liulab.dfci.harvard.edu/MACS/ for ChIP-seq MLRSeg Detection of NA In preparation. expressed regions from RNA-seq NextGeneId Transcript NA http://genome.crg.cat/software/geneid/index.html reconstruction PeakSeq Peak calling 19122651 http://www.gersteinlab.org/proj/PeakSeq/ for ChIP-seq 2 QuEST Peak calling 19160518 http://mendel.stanford.edu/sidowlab/downloads/quest/ for ChIP-seq RSEQTools RNASeq data NA http://rseqtools.gersteinlab.org processing Sole-search Peak calling 19906703 http://chipseq.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi for ChIP-seq SPP Peak calling 19029915 http://compbio.med.harvard.edu/Supplements/ChIP-seq for ChIP-seq ZINBA Peak calling In In preparation for FAIRE preparation (Zero Inflated Negative Binomial Algorithm)
C. Integration Tools and Resources Software Task PMID Availability ACT Aggregate NA http://act.gersteinlab.org signals over genes and other features AlignACE Motif Finder NA http://arep.med.harvard.edu/mrnadata/mrnasoft.html AnnoTrack Track manual 20923551 http://annotrack.sanger.ac.uk annotation & integrate external data BEDtools BED file set 20110278 http://code.google.com/p/bedtools/ operations and proximity analysis BindBoost Supervised NA Currently planned to be released in early 2011 learning framework for integrative models of TF binding CAGT Clustered NA http://code.google.com/p/cagt/
3 aggregation plots over TF binding sites and other genomic features CHAI Conservation 19286520 Code available on request. analysis ChromHMM Hidden NA In preparation. Markov Model segmentation of functional genomics data 18849524 Enredo-Pecan- All placental http://www.ebi.ac.uk/~jherrero/downloads/enredo/ + Ortheus mammals 18849525 http://www.ebi.ac.uk/~bjp/pecan/ whole http://www.ebi.ac.uk/~bjp/ortheus/ genome alignments Galaxy BED file set 16169926 http://bitbucket.org/galaxy/galaxy-central/wiki/GetGalaxy operations
Genomedata Genome data 20435580 http://noble.gs.washington.edu/proj/genomedata/ storage and access format. Genome Robust 17571346 http://www.encodestatistics.org/svn/genome_structural_correction/ Structure statistical test Correction for overlap significance. GERP v2.1 Conservation 15965027 http://mendel.stanford.edu/SidowLab/downloads/gerp/index.html analysis, per- base and features MEME Motif Finder 7584402 http://meme.sdsc.edu/meme4_4_0/intro.html MultiZ All 15060014 http://www.bx.psu.edu/miller_lab/ vertebrates
4 whole genome alignments PHAST Phylogenetic Submitted http://compgen.bscb.cornell.edu/phast/ alignment. R Statistical NA http://www.r-project.org/ computing framework. Segtools Plotting and NA http://noble.gs.washington.edu/proj/segtools/ Tabulating segmentation data Segway Dynamic In http://noble.gs.washington.edu/proj/segway/ Bayesian preparation network segmentation of multitrack
functional genomics data Weeder Motif Finder 15215380 http://159.149.109.9/modtools/
5