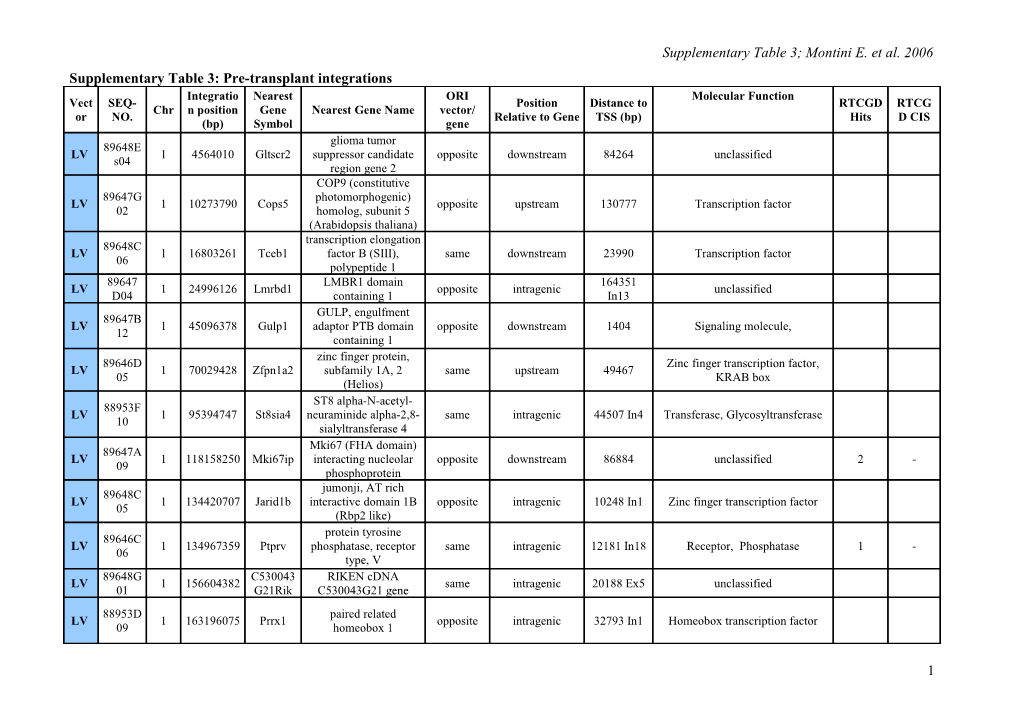
Supplementary Table 3; Montini E. et al. 2006 Supplementary Table 3: Pre-transplant integrations Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene glioma tumor 89648E LV 1 4564010 Gltscr2 suppressor candidate opposite downstream 84264 unclassified s04 region gene 2 COP9 (constitutive 89647G photomorphogenic) LV 1 10273790 Cops5 opposite upstream 130777 Transcription factor 02 homolog, subunit 5 (Arabidopsis thaliana) transcription elongation 89648C LV 1 16803261 Tceb1 factor B (SIII), same downstream 23990 Transcription factor 06 polypeptide 1 89647 LMBR1 domain 164351 LV 1 24996126 Lmrbd1 opposite intragenic unclassified D04 containing 1 In13 GULP, engulfment 89647B LV 1 45096378 Gulp1 adaptor PTB domain opposite downstream 1404 Signaling molecule, 12 containing 1 zinc finger protein, 89646D Zinc finger transcription factor, LV 1 70029428 Zfpn1a2 subfamily 1A, 2 same upstream 49467 05 KRAB box (Helios) ST8 alpha-N-acetyl- 88953F LV 1 95394747 St8sia4 neuraminide alpha-2,8- same intragenic 44507 In4 Transferase, Glycosyltransferase 10 sialyltransferase 4 Mki67 (FHA domain) 89647A LV 1 118158250 Mki67ip interacting nucleolar opposite downstream 86884 unclassified 2 - 09 phosphoprotein jumonji, AT rich 89648C LV 1 134420707 Jarid1b interactive domain 1B opposite intragenic 10248 In1 Zinc finger transcription factor 05 (Rbp2 like) protein tyrosine 89646C LV 1 134967359 Ptprv phosphatase, receptor same intragenic 12181 In18 Receptor, Phosphatase 1 - 06 type, V 89648G C530043 RIKEN cDNA LV 1 156604382 same intragenic 20188 Ex5 unclassified 01 G21Rik C530043G21 gene 88953D paired related LV 1 163196075 Prrx1 opposite intragenic 32793 In1 Homeobox transcription factor 09 homeobox 1
1 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89646C pre B-cell leukemia LV 1 168304346 Pbx1 opposite intragenic 62041 In2 Homeobox transcription factor 11 transcription factor 1 89646A LV 1 181065431 Nv1 nuclear VCP-like same intragenic 32542 In14 Hydrolase, Other hydrolase 11 88954A Sec61, alpha subunit 2 LV 2 590158 sec61a2 same upstream 88972 Membrane traffic protein 07 (S. cerevisiae) 88954C 4921504 RIKEN cDNA LV 2 20386169 opposite upstream 790248 unclassified 05 E06Rik 4921504E06 gene
88954F Signaling molecule, Peptide LV 2 62320400 Gcg glucagon opposite downstream 9122 02 hormone
longevity assurance 88953H LV 2 68794049 Lass6 homolog 6 (S. opposite intragenic 77091 In2 unclassified 05 cerevisiae) 89647D D430039 RIKEN cDNA LV 2 83527810 opposite intragenic 17332 In1 unclassified 11 N05Rik D430039N05 gene IMP1 inner 89647D mitochondrial LV 2 105621719 Immp1l same intragenic 12279 In1 Protease 10 membrane peptidase- like (S. cerevisiae) 88954A kinesin family member Cytoskeletal protein, Microtubule LV 2 108921037 Kif18a same intragenic 16511 In7 05 18A family DNA segment, Chr 2, 88953D D2Ertd7 LV 2 118336812 ERATO Doi 750, opposite intragenic 8959 In3 unclassified 08 50e expressed S- 89647A LV 2 154522054 Ahcy adenosylhomocysteine opposite intragenic 9310 In4 Hydrolase 04 hydrolase protein-L-isoaspartate 88953H 5330414 (D-aspartate) O- LV 2 181573553 same intragenic 854 Ex6 unclassified 07 D10Rik methyltransferase domain containing 2
2 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89647B Non-receptor serine/threonine LV 3 30414394 Prkci protein kinase C, iota same intragenic 7057 In1 01 protein kinase 89646H heat shock 70kDa Chaperone, Hsp 70 family LV 3 40172463 Hspa41 opposite intragenic 7152 In2 04 protein 4 like chaperone 89647G 3110057 RIKEN cDNA LV 3 40499797 opposite downstream 134266 unclassified 1 - 03 O12Rik 3110057O12 gene 89647E LV 3 41019407 Phf17 PHD finger protein 17 opposite intragenic 8842 In2 Zinc finger Transcription factor 1 - s01 89647A Nucleic acid binding, Nuclease, LV 3 54365626 Exosc8 exosome component 8 opposite intragenic 3988 In7 03 Exoribonuclease, Hydrolase spastic paraplegia 20, 89648H spartin (Troyer LV 3 54762765 Spg20 same intragenic 16288 In7 unclassified 10 syndrome) homolog (human) 89646B arginine/serine-rich LV 3 66858116 Rsrc1 same intragenic 200085 In5 unclassified 08 coiled-coil 1 89646B LPS-responsive beige- 519566 LV 3 86488645 Lrba same intragenic Membrane traffic protein 1 - 01 like anchor In38 89647H mannosidase, alpha, LV 3 100013792 Man1a2 opposite intragenic 97347 Ex10 Hydrolase 01 class 1A, member 2 N-deacetylase/N- 88954B sulfotransferase Protease, Cysteine protease, LV 3 124053682 Ndst4 opposite upstream 266005 09 (heparin glucosaminyl) Transferase 4 89648B 1810074 RIKEN cDNA LV 4 25406352 opposite intragenic 10945 In6 unclassified 09 P20Rik 1810074P20 gene RNA 89648C Phosphatase, Transferase, LV 4 33642365 Rngtt guanylyltransferase and opposite intragenic 53141 In9 1 - 10 Nucleotidyltransferase 5'-phosphatase 89646C leucine rich repeat Receptor, G-protein coupled LV 4 37585233 Lrrn6c opposite upstream 1600969 12 neuronal 6C receptor 88953B zinc finger, DHHC LV 4 81812088 Zdhhc21 same intragenic 33612 In6 unclassified 03 domain containing 21 hematopoietin/interferon-class 88953G interferon alpha family LV 4 87845453 Ifna1 opposite downstream 8605 (D200-domain) cytokine receptor 12 gene 1 binding
3 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene cytochrome P450, 88953E LV 4 95563963 Cyp2j5 family 2, subfamily j, same intragenic 75186 In1 Oxidoreductase, Oxygenase s07 polypeptide 5 89647H LV 4 108704627 Faf1 Fas-associated factor 1 opposite intragenic 69094 In6 Miscellaneous function 07 elongation of very long chain fatty acids 89647G LV 4 117381034 Elovl1 (FEN1/Elo2, same upstream 6013 Transferase, Acyltransferase 05 SUR4/Elo3, yeast)-like 1 splicing factor proline/glutamine rich Transcription factor, Nucleic acid 88954A LV 4 125976234 Sfpg (polypyrimidine tract opposite upstream 72215 binding, Ribonucleoprotein, Other 04 binding protein RNA-binding protein associated) tumor necrosis factor Receptor, Tumor necrosis factor 89647C LV 4 144088433 Tnfrsf1b receptor superfamily, same downstream 39185 receptor, Defense/immunity 03 member 1b protein 88954D MGC671 expressed sequence nucleic acid binding, LV 4 146272442 same downstream 42907 07 81 MGC67181 nucleus 88953A Sloan-Kettering viral LV 4 153721443 Ski same upstream 6560 Transcription factor 1 - 04 oncogene homolog 89647A cyclin-dependent Non-receptor serine/threonine LV 5 3307750 Cdk6 same upstream 42568 10 kinase 6 protein kinase 88953E ankyrin repeat and IBR LV 5 3743329 Ankib1 same intragenic 65602 In5 unclassified s10 domain containing 1 A kinase (PRKA) Cytoskeletal protein, Actin family 89646A LV 5 3953707 Akap9 anchor protein (yotiao) same intragenic 19411 In1 cytoskeletal protein, Non-motor 10 9 actin binding protein 88954G LV 5 16248016 Cd36 CD36 antigen same intragenic 8074 In3 Receptor, Cell adhesion molecule 03 serine/arginine-rich 89646F Non-receptor serine/threonine LV 5 21895109 Srpk2 protein specific kinase opposite downstream 72145 05 protein kinase 2 88953A AY6167 cDNA sequence, Oxidoreductase, Other LV 5 66810813 same intragenic 10153 In1 03 53 AY616753 oxidoreductase
4 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene CCR4-NOT 88954H LV 5 95153338 Cnot6l transcription complex, opposite intragenic 26268 In1 Transcription factor 03 subunit 6-like 89648B mitochondrial LV 5 95270180 Mrpl1 same intragenic 12086 In7 unclassified 05 ribosomal protein L1 89648A 3830405 RIKEN cDNA LV 5 99825520 same intragenic 16069 Ex9 unclassified 05 G04Rik 3830405G04 gene Nucleic acid binding, Synthase and 89648E 2'-5' oligoadenylate synthetase, Synthetase, LV 5 114022173 Oas12 opposite downstream 320 s11 synthetase-like 2 Transferase, Nucleotidyltransferase, proteasome (prosome, 89647D LV 5 122405522 Pmsd9 macropain) 26S same intragenic 14178 In4 Protease 05 subunit, non-ATPase, 9 89646E LV 5 139369690 Snx8 sorting nexin 8 opposite intragenic 15902 In1 unclassified s02 89648D 2700050 RIKEN cDNA LV 5 146400756 opposite intragenic 78371 In5 unclassified 10 F09Rik 2700050F09 gene 89648E 6330406I RIKEN cDNA LV 5 148321738 opposite upstream 28316 unclassified s03 15Rik 6330406I15 gene core 1 UDP- galactose:N- 89646C LV 6 7828258 C1galt1 acetylgalactosamine- opposite intragenic 6594 In1 Transferase, Glycosyltransferase 05 alpha-R beta 1,3- galactosyltransferase 88953D Extracellular matrix, Extracellular LV 6 31723283 Podx1 podocalyxin-like opposite upstream 82238 03 matrix glycoprotein 89648G zinc finger CCCH type, LV 6 38495346 Zc3hav1 same upstream 4533 unclassified 03 antiviral 1 NADH dehydrogenase 88953F LV 6 39732815 Ndufb2 (ubiquinone) 1 beta opposite intragenic 2087 In2 Oxidoreductase 07 subcomplex, 2 89648G thiamin LV 6 43743941 Tpk1 same intragenic 86799 In2 Kinase, Transferase 11 pyrophosphokinase LV 88954C 6 58806019 Abcg2 ATP-binding cassette, same intragenic 46872 In1 Transporter 04 sub-family G
5 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene
(WHITE), member 2 growth arrest and 89648H LV 6 67331373 Gadd45a DNA-damage- same upstream 41232 Miscellaneous function 09 inducible 45 alpha succinate-CoA ligase, 88954G LV 6 73595954 Suclg1 GDP-forming, alpha same intragenic 9583 In2 Synthase and synthetase 07 subunit 88954H Cell adhesion molecule, CAM LV 6 106397098 Cntn4 contactin 4 opposite intragenic 232328 In2 07 family adhesion molecule 89646D 2700091 RIKEN cDNA LV 6 120818908 same upstream 29538 unclassified 03 N06Rik 2700091N06 gene 88954D C-type lectin domain LV 6 123973920 Clec4e opposite upstream 16891 Receptor 04 family 4, member e 88954C LV 6 124890561 Cd165 CD163 antigen same upstream 26773 Receptor 06 killer cell lectin-like 88954D Receptor, Defense/immunity LV 6 130619068 Klra5 receptor, subfamily A, opposite intragenic 2139 In2 02 protein member 5 89646F inositol 1,4,5- 362789 Receptor, Ion channel, Ligand- LV 6 147059816 Itpr2 opposite intragenic 5 - 09 triphosphate receptor 2 In53 gated ion channell hect (homologous to the E6-AP (UBE3A) 89647E carboxyl terminus) 130687 Select regulatory molecule, G- LV 7 50450418 Herc2 opposite intragenic s07 domain and RCC1 In55 protein modulator (CHC1)-like domain (RLD) 2 88953A Receptor, G-protein coupled LV 7 98062196 Olfr633 olfactory receptor 633 opposite downstream 2334 12 receptor 89648H male sterility domain LV 7 107359656 Mlstd2 opposite upstream 6723 Oxidoreductase, Reductase 1 - 11 containing 2 88953E nucleosome assembly LV 7 137950031 Nap1l4 same intragenic 12079 In4 Chaperone s09 protein 1-like 4 tubulin, gamma Microtubule family cytoskeletal 89647G LV 8 12021693 Tubgcp3 complex associated opposite intragenic 29043 In11 protein, Non-motor microtubule 12 protein 3 binding protein
6 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 88953E LV 8 59859111 Clcn3 chloride channel 3 same downstream 98923 Anion channel s01 88954A 4732435 RIKEN cDNA LV 8 67262112 opposite downstream 243734 Transferase, Glycosyltransferase 2 - 10 N03Rik 4732435N03 gene 89648A 2410193 RIKEN cDNA Transporter, Transfer/carrier LV 8 77892187 opposite intragenic 169491 In8 1 - 06 C02Rik 2410193C02 gene protein, Mitochondrial 89648F inositol polyphosphate- LV 8 80734425 Inpp4b same upstream 214486 Phosphatase 02 4-phosphatase, type II CWF19-like 2, cell 89647H LV 9 3378516 Cwf1912 cycle control (S. same intragenic 70746 In16 unclassified 04 pombe) 89646G platelet-derived growth Receptor Signaling molecule, LV 9 6034412 Pdgfd opposite upstream 85103 05 factor, D polypeptide Growth factor Microtubule family cytoskeletal 89646G dynein, cytoplasmic, 189693 LV 9 6979570 Dnchc2 opposite intragenic protein, Microtubule binding 06 heavy chain 2 In57 motor protein, Hydrolase 88953E baculoviral IAP repeat- LV 9 7857864 Birc3 same intragenic 21128 In5 unclassified s08 containing 3
89648D Select regulatory molecule, G- LV 9 21363840 Dnm2 dynamin 2 same intragenic 50270 In7 12 protein, Small GTPase 89648G junction adhesion LV 9 27005018 Jam3 same intragenic 43353 In1 unclassified 1 - 12 molecule 3 89647B Rho GTPase-activating Select regulatory molecule, G- LV 9 31985614 Grit opposite upstream 143774 05 protein protein modulator 89647F D130054 RIKEN cDNA LV 9 45900900 same intragenic 19230 In6 unclassified 11 N24Rik D130054N24 gene 89646H RNA binding motif LV 9 48506087 Rbm7 same intragenic 6591 Ex5 Nucleic acid binding 12 protein 7 88953D Cytoskeletal protein, Actin family LV 9 72709362 Tex9 testis expressed gene 9 same upstream 65789 1 - 05 cytoskeletal protein 89647F phospholipid LV 9 92192794 Plscr2 same downstream 3786 Transfer/carrier protein 08 scramblase 2
7 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 88954B phospholipid LV 9 92221237 Plscr2 same downstream 32229 Transfer/carrier protein 12 scramblase 2 propionyl Coenzyme A 88953C LV 9 101194798 Pccb carboxylase, beta opposite upstream 258375 Ligase, Other ligase 10 polypeptide 89646G RNA binding motif Nucleic acid binding, Other RNA- LV 9 107891397 Rbm6 same intragenic 49289 In6 10 protein 6 binding protein 6-phosphofructo-2- 89646G Phosphatase, Carbohydrate LV 9 109080857 Pfkfb4 kinase/fructose-2,6- opposite downstream 2047 08 phosphatase biphosphatase 4 89648B expressed sequence LV 9 122048648 C85492 same upstream 32509 transferase activity 11 C85492 89648C 300538 Select calcium binding protein, LV 10 12442774 Utrn utrophin same intragenic 03 In51 Cytoskeletal protein 89647A interferon gamma Receptor, Cytokine receptor, LV 10 19452440 Ifngr1 opposite upstream 66819 02 receptor 1 Interferon receptor 89646E E230031 hypothetical protein Receptor, Cytokine receptor, LV 10 19673492 same intragenic 33012 In2 2 - s09 K19Rik E230031K19 Interleukin receptor solute carrier family 16 88953D (monocarboxylic acid LV 10 40216771 Slc16a10 same intragenic 2943 In1 Transporter 02 transporters), member 10 89646A 1700060 RIKEN cDNA LV 10 55819367 opposite upstream 537877 Transcription factor 04 H10Rik 1700060H10 gene Select calcium binding protein, 89648D calcium binding atopy- LV 10 59572966 Cbara1 same upstream 91722 Calmodulin related protein, 1 - 07 related autoantigen 1 Annexin 89646B transmembrane protein LV 10 68707119 Tmem26 opposite upstream 71953 unclassified 1 - 05 26 89646B A930033 RIKEN cDNA LV 10 69311275 opposite upstream 42835 unclassified 02 H14Rik A930033H14 gene 88953E LV 10 79888329 Rnf126 ring finger protein 126 opposite intragenic 1220 In1 unclassified s02 guanine nucleotide 89646F Select regulatory molecule, G- LV 10 81667950 Gna11 binding protein, alpha same intragenic 12878 In4 02 protein, Large G-protein 11 LV 88953B 10 89403169 Nr1h4 nuclear receptor opposite downstream 23660 Receptor, Transcription factor, 2 - 8 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene subfamily 1, group H, 07 Nuclear hormone receptor member 4 nuclear receptor 88953E Receptor, Transcription factor, LV 10 89455807 Nr1h4 subfamily 1, group H, opposite intragenic 23271 In3 2 - s05 Nuclear hormone receptor member 4 89648A E030041 RIKEN cDNA LV 10 89763520 same intragenic 47169 In11 unclassified 08 M21Rik E030041M21 gene 88953C apoptotic peptidase Select regulatory molecule, Other LV 10 91051137 Apaf1 opposite intragenic 2888 In2 11 activating factor 1 enzyme regulator 89647E 4930430 RIKEN cDNA LV 10 101279841 opposite upstream 801214 unclassified s02 F08Rik 4930430F08 gene 89646D Membrane traffic regulatory LV 10 108298442 Syt1 synaptotagmin I opposite downstream 86659 10 protein dual-specificity 89648A tyrosine-(Y)- Non-receptor serine/threonine LV 10 118355300 Dyrk2 same downstream 85442 11 phosphorylation protein kinase regulated kinase 2 88954B uridine phosphorylase LV 11 9510669 Upp1 opposite downstream 479712 Transferase, Phosphorylase 02 1 88953F LV 11 12574660 Cob1 cordon-bleu same upstream 215067 unclassified 03 89648D BC02712 cDNA sequence LV 11 14455131 opposite upstream 1697435 unclassified 02 7 BC027127 Nucleic acid binding, Nuclease, polyribonucleotide 88953A Exoribonuclease, Transferase, LV 11 29051946 Pnpt1 nucleotidyltransferase same intragenic 26404 In22 09 Nucleotidyltransferase, Hydrolase, 1 Esterase 89646B Musashi homolog 2 Nucleic acid binding, LV 11 88464673 Msi2h same upstream 146531 2 - 12 (Drosophila) Ribonucleoprotein COX11 homolog, Transporter, Cation transporter, 89647C cytochrome c oxidase LV 11 90493745 Cox11 opposite downstream 27722 Hydrogen transporter, 07 assembly protein Oxidoreductase, Oxidase (yeast) 88953E RAB5C, member RAS Select regulatory molecule, G- LV 11 100555097 Rab5c opposite intragenic 4181 In1 2 CIS s04 oncogene family protein
9 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89646F BC03086 cDNA sequence LV 11 102077046 opposite intragenic 7073 Ex3 unclassified 10 7 BC030867 pleckstrin homology 88953G domain containing, LV 11 103206486 Plekhm1 same intragenic 27268 In5 unclassified 1 - 08 family M (with RUN domain) member 1 89646H LV 11 119092029 Gaa glucosidase, alpha, acid opposite intragenic 2891 In2 Hydrolase, Glucosidase 11 89646A synaptophysin-like transporter activity, transport, LV 12 29565419 Syp1 opposite intragenic 5249 In1 05 protein synaptic vesicle 89648B syntaxin binding LV 12 41686863 Stxbp6 same intragenic 133545 In2 unclassified 02 protein 6 (amisyn) survivor of motor Nucleic acid binding, mRNA 89647B LV 12 55757412 Sip1 neuron protein same intragenic 10786 In7 processing factor, mRNA splicing 1 - 07 interacting protein 1 factor, Other RNA-binding protein meningioma expressed 89646A LV 12 55935939 Mgea6 antigen 6 (coiled-coil same downstream 12718 unclassified 06 proline-rich) 88953F gene model 527, LV 12 60894796 Gm527 same upstream 801055 unclassified 02 (NCBI) 88954E FERM domain LV 12 67663852 Frmd6 same intragenic 4556 In1 unclassified s02 containing 6 89648H LV 12 74813803 Gphn gephyrin same upstream 269531 Miscellaneous function 12 serine 89647G palmitoyltransferase, LV 12 84190830 Spt1c2 same intragenic 77099 In5 Transferase, Other transferase 11 long chain base subunit 2 serine 88953A palmitoyltransferase, LV 12 84235355 Spt1c2 same intragenic 32574 In3 Transferase, Other transferase 05 long chain base subunit 2 89647H 2610021 RIKEN cDNA LV 12 95101358 same downstream 43793 unclassified 1 - 02 K21Rik 2610021K21 gene LV 89646F 12 108429458 Brf1 BRF1 homolog, opposite downstream 6586 Transcription factor, Basal 01 subunit of RNA transcription factor polymerase III
10 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene transcription initiation factor IIIB (S. cerevisiae) guanosine diphosphate Select regulatory molecule, G- 89647F LV 13 3457838 Gdi2 (GDP) dissociation opposite downstream 7637 protein modulator, Transferase, 02 inhibitor 2 Acyltransferase 89647C pitrilysin LV 13 6470153 Pitrm1 same intragenic 25059 In20 Metalloprotease 05 metallepetidase 1 CDK5 regulatory 88953C LV 13 29333432 Cdkal1 subunit associated opposite intragenic 1779 In2 unclassified 12 protein 1-like 1 88954C orofacial cleft 1 LV 13 39489907 Ofcc1 same downstream 349236 unclassified 03 candidate 1
88954F jumonji, AT rich LV 13 44383379 Jarid2 opposite intragenic 77831 In2 Transcription factor 11 interactive domain 2
89646E dystrobrevin binding LV 13 44704430 Dtnbp1 same upstream 128387 unclassified s03 protein 1 MAK10 homolog, 89648D amino-acid N- LV 13 58262831 Mak10 same intragenic 17821 In6 unclassified 01 acetyltransferase subunit, (S. cerevisiae) hyaluronan and 88954E LV 13 85557412 Hapln1 proteoglycan link same upstream 98108 Extracellular matrix glycoprotein s01 protein 1 procollagen, type IV, 88954D alpha 3 (Goodpasture Non-receptor serine/threonine LV 13 92797954 Col4a3bp same intragenic 63881 In7 10 antigen) binding protein kinase protein poly (ADP-ribose) 89648F 144630 LV 13 113333080 Parp8 polymerase family, opposite intragenic unclassified 12 In13 member 8 88954D topoisomerase (DNA) Select regulatory molecule, LV 14 14893436 Top2b same intragenic 31613 In10 11 II beta Nucleic acid binding, Isomerase 88954A LV 14 46524222 V2r11/10 V2r11/10 opposite upstream 22164 08 11 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89646G LV 14 51119916 Cenpj centromere protein J same upstream 26157 unclassified 03 89646B gonadotropin releasing LV 14 62426555 Gnrh1 opposite downstream 148712 Peptide hormone 03 hormone 1 88954G 4921530 RIKEN cDNA LV 14 90434104 opposite downstream 82738 unclassified 01 L21Rik 4921530L21 gene 89647F endothelin receptor Receptor, G-protein coupled LV 14 98531352 Ednrb opposite upstream 142663 01 type B receptor
89646E Receptor, Cytokine receptor, LV 15 9543126 I17r interleukin 7 receptor opposite upstream 199783 s10 Defense/immunity protein 89646A 9230109 RIKEN cDNA LV 15 25205218 opposite upstream 66911 unclassified 1 - 07 A22Rik 9230109A22 gene Transporter, Transfer/carrier 89646E LV 15 27490988 Ank progressive ankylosis same intragenic 34980 In1 protein, Mitochondrial carrier s08 protein tyrosine 3- monooxygenase/trypto 89648E LV 15 36773176 Ywhaz phan 5-monooxygenase same downstream 488 miscellaneous function protein s09 activation protein, zeta polypeptide 88953H trichorhinophalangeal Transcription factor, Nucleic acid LV 15 48930088 Trps1 opposite downstream 1729198 09 syndrome I (human) binding, Nuclease eukaryotic translation 89647A Nucleic acid binding, Translation LV 15 51352218 Eif3s3 initiation factor 3, opposite downstream 431576 06 factor, Other RNA-binding protein subunit 3 (gamma) eukaryotic translation 88954C Nucleic acid binding, Translation LV 15 51838321 Eif3s3 initiation factor 3, opposite intragenic 24370 In2 02 factor, Other RNA-binding protein subunit 3 (gamma) 89646G DEP domain G-protein modulator, Guanyl- LV 15 55167885 Depdc6 same intragenic 53604 In3 11 containing 6 nucleotide exchange factor KH domain containing, 88953A RNA binding, signal Nucleic acid binding, Other RNA- LV 15 70276747 Khdrbs3 opposite downstream 1158694 10 transduction associated binding protein 3
12 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 88953F IMP dehydrogenase/GMP LV 16 15090356 Dnm1l dynamin 1-like opposite intragenic 36410 In12 06 reductase 88954D IMP dehydrogenase/GMP LV 16 15112293 Dnm1l dynamin 1-like opposite intragenic 14473 In2 08 reductase 89647C transmembrane serine LV 16 44560574 Tmprss7 same intragenic 8942 In7 Protease, Serine protease 01 protease 7 89646D LV 16 56687920 Col8a1 opposite upstream 146603 01 88953G 5-hydroxytryptamine Receptor, G-protein coupled LV 16 64074417 Htr1f opposite intragenic 57958 In2 1 - 02 (serotonin) receptor 1F receptor 88953A immunoglobulin LV 16 66626521 Igsf4d opposite intragenic 6802 In1 Receptor 01 superfamily, member 4 Transcription factor, Nucleic acid 89648E chromodomain helicase LV 17 13757874 Chd1 same intragenic 49046 In23 binding, Helicase, DNA helicase, s07 DNA binding protein 1 Hydrolase 89646A LV 17 22546344 Sepx1 selenoprotein X 1 same intragenic 4585 In3 Oxidoreductase, Reductase 02 89648D testis expressed gene Miscellaneous function, Other LV 17 27894131 Tex27 same intragenic 79003 In2 1 - 11 27 miscellaneous function protein 89648C Receptor miscellaneous function LV 17 43732773 Xpo5 exportin 5 opposite intragenic 19996 In14 11 protein 89648H LV 17 43898757 Zfp318 zinc finger protein 318 same intragenic 3812 In1 unclassified 04 fer (fms/fps related) 89646H Kinase, Protein kinase, Non- LV 17 61678598 Fert2 protein kinase, testis opposite intragenic 40637 In4 08 receptor tyrosine protein kinase specific 2 89647E Transcription factor, Nucleic acid LV 17 86320591 Foxn2 forkhead box N2 same intragenic 18255 In1 s06 binding 89647E follicle stimulating Receptor, G-protein coupled LV 17 86908410 Fshr opposite intragenic 155158 In3 s09 hormone receptor receptor Transcription factor, Zinc finger 89646C zinc finger homeobox LV 18 5568437 Zfhx1a same upstream 28199 transcription factor, Homeobox 07 1a transcription factor
13 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89647D tumor necrosis factor, LV 18 50106921 Tnfaip8 opposite upstream 161696 unclassified 1 - 02 alpha-induced protein 8 89647B Transcription factor, Basic helix- LV 18 69830552 Tcf4 transcription factor 4 opposite intragenic 255546 In8 1 - 03 loop-helix 89648D MAD homolog 4 LV 18 73815184 Smad4 same downstream 58060 Transcription factor 1 - 05 (Drosophila) 88953G MAD homolog 4 LV 18 73891367 Smad4 same intragenic 46604 In7 Transcription factor 01 (Drosophila) solute carrier family 14 88954F LV 18 78642021 Slc14a2 (urea transporter), opposite intragenic 77596 In2 Transporter 1 - 10 member 2 89646B ATPas, class II, type Transporter, Cation transporter LV 18 81000480 Atp9b opposite intragenic 56931 In6 2 - 09 9B Hydrolase 88953B HRAS like suppressor LV 19 7286123 Hrasls3 opposite intragenic 14754 In2 Select regulatory molecule 01 3 89648F ciliary neurotrophic Signaling molecule, Growth factor, LV 19 11973240 Cntf opposite intragenic 1773 In3 01 factor Neurotrophic factor 89646B 5730446 RIKEN cDNA LV 19 20915399 same intragenic 24162 In1 unclassified 06 C15Rik 5730446C15 gene RNA terminal 89648C Nucleic acid binding, Other RNA- LV 19 28396007 Rcl1 phosphate cyclase-like same intragenic 42032 Ex9 02 binding protein, Lyase, Cyclase 1 88953G 1700095 RIKEN cDNA LV 19 33531062 opposite intragenic 7684 In1 unclassified 06 N21Rik 1700095N21 gene 89647C LV Un 28093905 H2-B1 opposite upstream 16924 08 ubiquitously 89647E transcribed LV X 16438817 Utx opposite intragenic 37209 In3 Miscellaneous function s10 tetratricopeptide repeat gene, X chromosome 89648H cytosolic ovarian LV X 43475103 Cova1 same downstream 56216 unclassified 03 carcinoma antigen 1 solute carrier family 9 89646D LV X 51418378 Slc9a6 (sodium/hydrogen same downstream 1770 Transporter, Cation transporter 12 exchanger), isoform 6
14 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89647G methyl CpG binding LV X 68709054 Mecp2 opposite intragenic 37195 In2 Nucleic acid binding 07 protein 2 pyruvate 89648H LV X 88510341 Pdk3 dehydrogenase kinase, same upstream 17587 Kinase, Protein kinase 1 - 06 isoenzyme 3 Microtubule family cytoskeletal 89648E kinesin family member LV X 95326393 Kif4 opposite intragenic 99294 In28 protein, Microtubule binding s12 4 motor protein 89648E cellular nucleic acid LV X 98202727 Cnbp2 same upstream 29225 Nucleic acid binding s08 binding protein 2 88953B ribosomal protein S6 121365 Non-receptor serine/threonine LV X 105816066 Rps6ka6 same intragenic 08 kinase polypeptide 6 In12 protein kinase Nucleic acid binding, Nuclease, Endoribonuclease, Cytoskeletal 88954G diaphanous homolog 2 LV X 123811936 Diap2 opposite downstream 23948 protein, Actin family cytoskeletal 05 (Drosophila) protein, Non-motor actin binding protein, Hydrolase, Esterase 89648F LV X 138978222 Amot angiomotin opposite upstream 51129 unclassified 05 89646E LV X 149221176 Prdx4 peroxiredoxin 4 opposite upstream 256572 Oxidoreductase s11 89647B cytidine 5'-triphosphate 115729 Synthase and synthetase, Synthase, LV X 156616740 Ctps2 same intragenic 08 synthase 2 In15 Ligase Microtubule family cytoskeletal 89646E LV X 163617585 Mid1 midline 1 same downstream 16156 protein, Non-motor microtubule 4 CIS s01 binding protein 89648E LV LINE, L1_Mus1 s01 89648E LV Satellite GSAT_MM s02 89647E LTR, ERVL, LV s05 MERVL_Mm 89648E LV Satellite GSAT_MM s05 89648E LV Satellite GSAT_MM s06
15 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 88954E LV multiple hits s08 88953E LV Satellite GSAT_MM s11 88953B LV SINE, B2_Mm2 02 88953D LV Satellite GSAT_MM 01 88953H LV Satellite GSAT_MM 01 88953H LV LINE, L1_Mm 06 88953H LV simple repeat 08 88954B LV multiple hits 07 88954H LV LINE, L1_Mus1 02 89646A LV satellite GSAT_MM 08 89646A LV satellite GSAT_MM 09 89646B LV satellite GSAT_MM 07 89646C LV satellite GSAT_MM 03 89646C LV satellite GSAT_MM 04 89646F LV satellite GSAT_MM 03 89646F LV satellite GSAT_MM 06 89646G LV satellite GSAT_MM 02 89646G LV satellite GSAT_MM 04 LV 89646G satellite GSAT_MM
16 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 09 89646H LV satellite GSAT_MM 02 89646H LV satellite GSAT_MM 10 89647A LV Satellite GSAT_MM 01 89647A LV Satellite GSAT_MM 11 89647A LV Satellite GSAT_MM 12 89647B LV Satellite GSAT_MM 02 89647B LV Satellite GSAT_MM 10 89647B LV Satellite GSAT_MM 11 89647C LV Satellite GSAT_MM 10 89647C LV SINE, B1_Mus2, Alu 12 89647D LV Satellite GSAT_MM 03 89647D LV Satellite GSAT_MM 09 89647F LV simple repeat 05 89647H LV Satellite and LTR 12 89648A LV Satellite GSAT_MM 10 89648B LV Satellite GSAT_MM 03 89648B LV Satellite GSAT_MM 07 89648B LV LTR, ERVK 10
17 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89648C LV Satellite GSAT_MM 04 89648C LV LINE 08 89648F LV Satellite GSAT_MM 03 89648F LV Satellite GSAT_MM 06 89648F LV Satellite GSAT_MM 07 89648F LV LINE, L1, Lx7 09 89648F LV Satellite GSAT_MM 11 89648H LV Satellite GSAT_MM 07 amyotrophic lateral 89755D sclerosis 2 (juvenile) RV 1 58983863 Als2cr12 opposite intragenic 16963 In5 unclassified 05 chromosome region, candidate 12 (human)
89792A interleukin 8 receptor, RV 1 74321576 Il8rb opposite upstream 13747 G-protein coupled receptor 1 - 03 beta
protein kinase, AMP- 89791G Non-receptor serine/threonine RV 1 75050048 Prkag3 activated, gamma 3 opposite intragenic 261 In1 11 protein kinase non-catatlytic subunit
89791A integral membrane Membrane-bound signaling RV 1 85703004 Itm2c opposite intragenic 2082 In1 1 - 08 protein 2C molecule 89792A 4930438 RIKEN cDNA RV 1 86033810 same intragenic 72706 In19 12 O05 4930438O05 gene RV 89754F 1 87777729 Ugt1a10 UDP same upstream 96829 Glycosyltransferase 05 glycosyltransferase 1 family, polypeptide
18 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene
A10
89754G glutamine repeat RV 1 88399118 Glrp1 opposite upstream 69946 unclassified 06 protein 1 peptidylglycine alpha- 89754D RV 1 97858715 Pam amidating same intragenic 16750 In1 Oxidoreductase, Oxygenase 07 monooxygenase tumor necrosis factor 89792B Tnfrsf11 Cytokine receptor, Tumor necrosis RV 1 105824464 receptor superfamily, opposite downstream 37930 2 CIS 02 a factor receptor member 11a
89792D receptor interacting RV 1 132274053 Ripk5 same upstream 846 unclassified 03 protein kinase 5 89792G A130050 RIKEN cDNA RV 1 137658247 opposite upstream 120827 unclassified 2 - 08 O07Rik A130050O07 gene phospholipase A2, 89791E RV 1 149783193 Pla2g4a group IVA (cytosolic, opposite intragenic 14819 In1 Hydrolase, Lipase, Phospholipase s01 calcium-dependent) 89792C sterol O-acyltransferase RV 1 156314692 Soat1 same downstream 21904 Transferase, Acyltransferase 08 1 89791H immunoglobulin RV 1 172240542 Igsf8 opposite upstream 785 miscellaneous function protein 08 superfamily, member 8 glyceraldehyde-3- 89754A MGC683 phosphate RV 1 180385747 opposite upstream 100950 Oxidoreductase, Dehydrogenase 1 - 02 23 dehydrogenase (phosphorylating)-like 89792B TRAF3 interacting RV 1 192939755 Traf3ip3 same intragenic 494 Ex2 unclassified 06 protein 3 89755F FERM domain 407028 RV 2 4477172 Frmd4a opposite intragenic unclassified 1 - 07 containing 4A In12 calcium channel, 89755C RV 2 14558437 Cacnb2 voltage-dependent, same intragenic 27858 In2 Voltage-gated calcium channel 05 beta 2 subunit RV 89755F 2 14887731 Cacnb2 calcium channel, same intragenic 357152 In5 Voltage-gated calcium channel 02 voltage-dependent,
19 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene
beta 2 subunit Membrane-bound signaling 89791E Notch gene homolog 1 RV 2 26462556 Notch1 same upstream 26756 molecule, Defense/immunity 11 CIS s08 (Drosophila) protein pleckstrin homology, 89791D Sec7 and coiled-coil RV 2 58081092 Pscdbp opposite intragenic 4507 In1 unclassified 11 domains, binding protein ubiquitin-conjugating 89755C enzyme E2E 3, RV 2 78776966 Ube2e3 same downstream 158634 Ligase, Other ligase 11 UBC4/5 homolog (yeast) cAMP responsive 89755E Transcription factor, Nucleic acid RV 2 91760946 Creb3l1 element binding same upstream 32065 s12 binding, Nuclease protein 3-like 1 89754 B230118 RIKEN cDNA RV 2 101289493 same intragenic 44177 In4 unclassified B06 H07Rik B230118H07 gene 89791B Arhgap1 Rho GTPase activating RV 2 113471060 opposite intragenic 794 In1 unclassified 07 1a protein 11A 89755C AA4671 expressed sequence RV 2 122166641 opposite downstream 11711 Oxidoreductase 1 - 03 97 AA467197 sema domain, transmembrane domain 89754D Membrane-bound signaling RV 2 124217809 Sema6d (TM), and cytoplasmic same downstream 38646 11 molecule domain, (semaphorin) 6D solute carrier family 27 89755D RV 2 126013429 Slc27a2 (fatty acid transporter), same upstream 53580 Transporter, Ligase, Other ligase 10 member 2 Select regulatory molecule, 89791C cystatin 10 RV 2 148738876 Cst10 same upstream 123310 Protease inhibitor, Cysteine 10 (chondrocytes) protease inhibitor 89791C tribbles homolog 3 Non-receptor serine/threonine RV 2 151807820 Trib3 opposite upstream 6851 12 (Drosophila) protein kinase 89791A RV 2 154071481 Zfp341 zinc finger protein 341 opposite intragenic 1175 In1 unclassified 06
20 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene protein phosphatase 1, 89755H Transcription factor, Nucleic acid RV 2 158148818 Ppp1r16b regulatory (inhibitor) same intragenic 26450 In2 01 binding subunit 16B UDP-Gal:betaGlcNAc 89755E beta 1,4- RV 2 166823438 B4galt5 same upstream 17521 Transferase, Glycosyltransferase 1 - s07 galactosyltransferase, polypeptide 5 transmembrane, 89754F RV 2 172798035 Tmepai prostate androgen same upstream 61034 unclassified 1 - 01 induced RNA solute carrier organic 89791G RV 2 180156077 Slco4a1 anion transporter same upstream 21860 Transporter 1 - 10 family, member 4a1 DnaJ (Hsp40) 89755H RV 2 181237021 Dnajc5 homolog, subfamily C, opposite upstream 442 Membrane traffic protein 12 member 5 89754F muscleblind-like 1 Nucleic acid binding, Other RNA- RV 3 60165837 Mbnl1 opposite upstream 13889 1 - 11 (Drosophila) binding protein homeodomain 89791B Non-receptor serine/threonine RV 3 103214648 Hipk1 interacting protein opposite intragenic 3923 In1 01 protein kinase kinase 1 proteasome (prosome, 89791H RV 3 108053909 Psma5 macropain) subunit, opposite intragenic 955 In1 Protease 02 alpha type 5 protein phosphatase 3, Protein phosphatase, Select 89755A RV 3 135706732 Ppp3ca catalytic subunit, alpha opposite intragenic 151076 In2 calcium binding protein, Other 1 - 03 isoform select calcium binding proteins EGF, latrophilin seven 89755B RV 3 149589391 Eltd1 transmembrane domain opposite upstream 828190 G-protein coupled receptor 08 containing 1 myeloid/lymphoid or mixed lineage- 89791F RV 4 86788862 Mllt3 leukemia translocation same intragenic 3645 In1 Transcription factor 1 - 04 to 3 homolog (Drosophila) 89754G kelch-like 9 RV 4 87692164 Klhl9 opposite downstream 16490 unclassified 02 (Drosophila)
21 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89791D cyclin-dependent RV 4 88453689 Cdkn2a same upstream 372597 unclassified 12 kinase inhibitor 2A 89792A 2310009 RIKEN cDNA RV 4 94532709 opposite upstream 21867 Carbohydrate kinase 1 - 10 E04Rik 2310009E04 gene 89791D acyl-CoA thioesterase RV 4 105746145 Acot11 same intragenic 12527 In1 Hydrolase, Esterase 06 11 erythroblast 89754G Zinc finger transcription factor, RV 4 118134386 Ermap membrane-associated opposite downstream 111 10 RING finger transcription factor protein 89791C eukaryotic translation Nucleic acid binding, Translation RV 4 125457432 Eif2c3 opposite upstream 924 05 initiation factor 2C, 3 initiation factor protein binding, cytoplasm, 89754D RV 4 133412216 Stmn1 stathmin 1 same upstream 17099 microtubule, intracellular signaling 04 cascade 89755B cell division cycle 42 Select regulatory molecule, G- RV 4 136204091 Cdc42 opposite intragenic 34833 In1 01 homolog (S. cerevisiae) protein, Small GTPase 89754E deleted in RV 4 149187454 Dnb5 same upstream 42917 Transporter 1 - s03 neuroblastoma 5 89755H cyclin-dependent Non-receptor serine/threonine RV 5 3369191 Cdk6 same intragenic 18873 In1 09 kinase 6 protein kinase protein tyrosine 89754F RV 5 19395035 Ptpn12 phosphatase, non- opposite downstream 44182 Protein phosphatase 1 - 08 receptor type 12 protein tyrosine 89754A RV 5 19403525 Ptpn12 phosphatase, non- opposite downstream 35692 Protein phosphatase 1 - 11 receptor type 12 89754C prolactin regulatory RV 5 29420087 Preb opposite upstream 913 Nucleic acid binding 1 - 12 element binding 89754C actin related protein 2/3 RV 5 121552209 Arpc3 opposite intragenic 7930 In2 unclassified 07 complex, subunit 3 89754A B-cell CLL/lymphoma RV 5 134182820 Bcl7b opposite intragenic 835 In1 unclassified 09 7B 89755C Homeobox transcription factor, RV 5 135455291 Cutl1 cut-like 1 (Drosophila) opposite intragenic 125450 In2 4 CIS 01 Nucleic acid binding 89754D Homeobox transcription factor, RV 5 135476435 Cutl1 cut-like 1 (Drosophila) opposite intragenic 104306 In2 3 CIS 08 Nucleic acid binding
22 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89754E 1110015 RIKEN cDNA RV 5 138774863 same upstream 2885 unclassified s01 K06Rik 1110015K06 gene 89754G Transcription factor, Nucleic acid RV 5 141381855 Foxk1 forkhead box K1 same upstream 25659 4 - 11 binding 89755A 9030611 RIKEN cDNA RV 6 65785974 opposite upstream 62449 unclassified 12 K07Rik 9030611K07 gene DNA segment, Chr 6, 89792A RV 6 83358423 D6Mm5e Miriam Meisler 5, opposite intragenic 58324 In6 unclassified 2 CIS 04 expressed 89755E 2300003 RIKEN cDNA RV 6 87019052 same upstream 44389 unclassified s04 P22Rik 2300003P22 gene 89792C AB04155 hypothetical protein, RV 6 88176813 opposite downstream 6623 unclassified 07 0 MNCb-4779 89755H 4933428 RIKEN cDNA Actin family cytoskeletal protein, RV 6 89018295 opposite downstream 34520 04 M03Rik 4933428M03 gene Non-motor actin binding protein microphthalmia- Basic helix-loop-helix 89755D RV 6 98286056 Mitf associated transcription same upstream 170970 transcription factor, Nucleic acid 07 factor binding 89792G RV 6 116337522 Wdr10 WD repeat domain 10 same intragenic 46456 In15 unclassified 09 Zinc finger transcription factor, 89755A C130073 RIKEN cDNA RV 6 125664726 opposite upstream 1084 KRAB box transcription factor, 06 D16Rik C130073D16 gene Nucleic acid binding killer cell lectin-like receptor activity, protein binding, 89755F RV 6 129545170 Klrb1b receptor subfamily B opposite upstream 28691 sugar binding, external side of 06 member 1D plasma membrane Receptor, Defense/immunity 89755F A630024 RIKEN cDNA RV 6 129716536 same upstream 16985 protein, Other defense and 4 CIS 03 B12Rik A630024B12 gene immunity protein Receptor, Signaling molecule, Membrane-bound signaling 89792C C-type lectin domain RV 6 129919610 Clec2d opposite downstream 51366 molecule, Defense/immunity 05 family 2, member d protein, Other defense and immunity protein Protein phosphatase, Select 89754D dual specificity RV 6 135510163 Dusp16 same intragenic 39781 In3 regulatory molecule, Kinase 06 phosphatase 16 inhibitor
23 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89755D 2310022 RIKEN cDNA RV 7 22950454 same intragenic 21543 In7 unclassified 1 - 03 A10Rik 2310022A10 gene Cysteine protease, Select calcium 89754D RV 7 25607616 Capns1 calpain, small subunit 1 same intragenic 432 In1 binding protein, Calmodulin 02 related protein 89791H Receptor, G-protein coupled RV 7 30516606 Olfr476 olfactory receptor 476 opposite upstream 55459 05 receptor 89791E AKT1 substrate 1 RV 7 38931165 Akt1s1 opposite downstream 828 unclassified s02 (proline-rich) 89754B AI48055 expressed sequence RV 7 39093393 same upstream 601 unclassified 09 6 AI480556 89754D 2410127 RIKEN cDNA Receptor, Nucleic acid binding, RV 7 39417462 opposite intragenic 20903 In24 09 E16Rik 2410127E16 gene Nuclease 89754G glycogen synthase 3, Synthase and synthetase, Synthase, RV 7 39537144 Gys3 same downstream 3354 09 brain Transferase, Other transferase 89792D glycogen synthase 3, Synthase and synthetase, Synthase, RV 7 39537335 Gys3 same downstream 3545 01 brain Transferase 89755D ATPase, class V, type RV 7 52952308 Atp10a opposite intragenic 15731 In1 Transporter, Hydrolase 08 10A 89791F RGM domain family, RV 7 67471885 Rgma same downstream 158592 unclassified 02 member A 89754A FCH and double SH3 RV 7 95200085 Fchsd2 same upstream 15287 unclassified 1 - 06 domains 2 89792C RV 7 96171808 Rnf121 ring finger protein 121 same upstream 717 unclassified 04 89755E RV 7 99186831 Olfr678 olfactory receptor 678 same downstream 4064 G-protein coupled receptor s05 89791H RV 7 104129215 Swap70 SWA-70 protein same intragenic 54582 In11 Nucleic acid binding 3 CIS 09 89791G RV 7 122109019 Itgam integrin alpha M opposite upstream 3498 Cell adhesion molecule 07 89754H regulator of G-protein G-protein modulator, Other G- RV 7 122441660 Rgs10 opposite intragenic 25892 In3 1 - 08 signalling 10 protein modulator zinc finger, RAN- 89754A RV 7 127298302 Zranb1 binding domain opposite upstream 17652 Nucleic acid binding, Nuclease 03 containing 1
24 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene protein tyrosine 89791B RV 7 129964314 Ptpre phosphatase, receptor opposite intragenic 58897 In1 Receptor, , Protein phosphatase 2 CIS 08 type, E 89791G Membrane-bound signaling RV 7 137417706 Tspan32 tetraspanin 32 same upstream 1005 05 molecule 89755B solute carrier family RV 8 5844048 Slc10a2 same upstream 1376278 Transporter, Cation transporter 12 10, member 2 DNA segment, Chr 8, 89792B D8Ertd8 RV 8 34979656 ERATO Doi 82, opposite downstream 29039 unclassified 1 - 09 2e expressed DNA segment, Chr 8, 89792B D8Ertd5 RV 8 46915147 ERATO Doi 594, same intragenic 45506 In1 unclassified 11 94e expressed LSM4 homolog, U6 Nucleic acid binding, mRNA 89755C small nuclear RNA RV 8 69831391 Lsm4 same intragenic 1650 In1 processing factor, mRNA splicing 1 - 07 associated (S. factor cerevisiae) solute carrier family 27 89792F RV 8 70729048 Slc27a1 (fatty acid transporter), same intragenic 10806 In3 Transporter, Ligase 3 CIS 11 member 1 89755C inositol polyphosphate- RV 8 81001194 Inpp4b same intragenic 52283 In4 Phosphatase 02 4-phosphatase, type II 89754B 4933431 RIKEN cDNA Select regulatory molecule, G- RV 8 82499371 opposite intragenic 86461 In7 01 N12Rik 4933431N12 gene protein modulator 89792F mannose receptor-like RV 8 110359017 Mrcl same intragenic 20790 In16 Protease, Hydrolase 07 precursor nudix (nucleoside 89755D RV 8 113430865 Nudt7 diphosphate linked opposite upstream 2203 Phosphatase, Hydrolase 04 moiety X)-type motif 7 89754E phospholipase C, Select calcium binding protein, RV 8 116875923 Plcg2 opposite intragenic 43081 In2 s08 gamma 2 Hydrolase, Lipase, Phospholipase 89791A coactosin-like 1 RV 8 119182123 Cotl1 same intragenic 15084 In2 unclassified 2 CIS 05 (Dictyostelium) 89792E BC02161 cDNA sequence RV 8 122147840 opposite upstream 14636 Synthase and synthetase s11 1 BC021611
25 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89754H 2310079 RIKEN cDNA RV 8 125029724 opposite intragenic 7965 In3 unclassified 01 N02Rik 2310079N02 gene 89791F ribosomal protein, RV 9 61786963 Rplp1 same downstream 246203 1 - 11 large, P1 acidic (leucine-rich) Select regulatory molecule, 89754B RV 9 62472043 Anp32a nuclear phosphoprotein same intragenic 10578 In1 Phosphatase modulator, 1 - 05 32 family, member A Phosphatase inhibitor 89754F MAD homolog 6 RV 9 64170353 Smad6 opposite upstream 28050 Transcription factor 06 (Drosophila) 89791H A230098 RIKEN cDNA RV 9 72943489 same upstream 31581 CAM family adhesion molecule 04 A12Rik A230098A12 gene 89754H coiled-coil domain RV 9 110706338 Ccdc12 opposite intragenic 8537 In1 unclassified 2 - 09 containing 12 89791F 2310001 RIKEN cDNA RV 9 121328856 opposite intragenic 14831 In1 unclassified 06 H13Rik 2310001H13 gene 89754E zinc finger CCCH-type RV 10 7707705 Zc3h12d opposite intragenic 9749 In2 unclassified s02 containing 12D 89755C E130306 RIKEN cDNA RV 10 9826879 same upstream 278365 unclassified 10 M17Rik E130306M17 gene solute carrier family 16 89792F (monocarboxylic acid RV 10 40240080 Slc16a10 same upstream 20366 Transporter 08 transporters), member 10 89791F popeye domain RV 10 45356283 Popdc3 same upstream 51378 unclassified 08 containing 3 89754A 2510003 RIKEN cDNA RV 10 62507121 opposite downstream 17737 unclassified 10 E04Rik 2510003E04 gene 89755B strawberry notch Nucleic acid binding, Nuclease, RV 10 80226774 Stno opposite upstream 860 11 homolog (Drosophila) Helicase B-cell translocation 89755E RV 10 96372295 Btg1 gene 1, anti- opposite upstream 222359 Miscellaneous function 1 - s06 proliferative Basic helix-loop-helix 89792H RV 10 107835095 Myf6 myogenic factor 6 opposite upstream 453295 transcription factor, Nucleic acid 01 binding RV 89754B 10 114940340 Tbc1d15 TBC1 domain family, opposite upstream 3143 unclassified
26 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 10 member 15 89754B RV 11 3175391 Zfp278 zinc finger protein 278 same upstream 9854 unclassified 1 - 08 transforming growth 89791E Select regulatory molecule, Kinase RV 11 6521388 Tbrg4 factor beta regulated opposite upstream 576 1 - s11 modulator gene 4 zinc finger protein, 89791H Zinc finger transcription factor, RV 11 11552720 Zfpn1a1 subfamily 1A, 1 same upstream 28286 1 - 01 KRAB box transcription factor (Ikaros) zinc finger protein, 89792F Zinc finger transcription factor, RV 11 11658113 Zfpn1a1 subfamily 1A, 1 same intragenic 77107 In7 1 - 04 KRAB box transcription factor (Ikaros) Zinc finger transcription factor, 89755C RV 11 51559017 Phf15 PHD finger protein 15 same downstream 10722 Other zinc finger transcription 06 factor, Nucleic acid binding 89792B spectrin domain with RV 11 61874909 Specc1 opposite intragenic 72169 In6 Cytoskeletal protein 08 coiled-coils 1 89754E 1110001 RIKEN cDNA RV 11 67555053 same downstream 13397 Oxidoreductase, Dehydrogenase s04 P11Rik 1110001P11 gene 89792C phospholipid Transfer/carrier protein, Other RV 11 69569114 Plscr3 opposite upstream 3221 1 - 10 scramblase 3 transfer/carrier protein 89754H active BCR-related RV 11 76208701 Abr same intragenic 26377 In2 G-protein modulator 12 gene 89755D kinase suppressor of 124560 Non-receptor serine/threonine RV 11 78747454 Ksr opposite intragenic 09 ras In13 protein kinase 89755F chaperonin subunit 6b RV 11 82489896 Cct6b same intragenic 14070 In5 Chaperone, Chaperonin 08 (zeta) 89754E Musashi homolog 2 Nucleic acid binding, RV 11 88279872 Msi2h same intragenic 38270 In1 2 - s10 (Drosophila) Ribonucleoprotein Receptor, Transcription factor, 89791G retinoic acid receptor, RV 11 98768790 Rara same intragenic 9842 In1 Nuclear hormone receptor, Nucleic 12 alpha acid binding 89754E RV 11 101358235 Brca1 breast cancer 1 same intragenic 11973 In2 Zinc finger transcription factor s11 89791B Receptor, Cell adhesion molecule, RV 11 104434576 Itgb3 integrin beta 3 same intragenic 5430 In1 11 Extracellular matrix glycoprotein
27 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89791A AK1293 cDNA sequence RV 11 120226848 opposite intragenic 31914 In10 unclassified 11 75 AK129375 89754D prolyl 4-hydroxylase, RV 11 120395110 P4hb same upstream 1146 Isomerase 10 beta polypeptide 89792D Transmembrane receptor RV 12 69205067 Rtn1 reticulon 1 opposite intragenic 47362 In1 12 regulatory/adaptor protein Transcription factor, Other 89792H Jun dimerization RV 12 82500451 Jundm2 same intragenic 31471 In2 transcription factor, Nucleic acid 8 CIS 11 protein 2 binding 89791D heat shock protein 1, RV 12 106172775 Hspca same upstream 1146 Chaperone 10 alpha RIKEN cDNA 89755G 2310040 RV 12 106172775 2310040A13 gene opposite upstream 40408 binding 10 A13 Wdr20 89754E Transcription cofactor, Nucleic RV 13 5600845 Klf6 Kruppel-like factor 6 same upstream 148225 s09 acid binding 89754G RV 13 13293985 Cts3 cathepsin 3 opposite upstream 916207 Protease, Cysteine protease 08 MRS2-like, 89792D magnesium DNA binding, membrane, metal RV 13 24500925 Mrs21 same upstream 668 11 homeostasis factor (S. ion transport cerevisiae) Select calcium binding protein, 89755F coagulation factor XIII, Calmodulin related protein, RV 13 36597140 F13a1 opposite upstream 6974 05 A1 subunit Annexin, Transferase, Acyltransferase Defense/immunity protein, 89791C AW4568 expressed sequence RV 13 48145596 same downstream 30336 Immunoglobulin receptor family 1 - 03 74 AW456874 member 89791E dopamine receptor Receptor, G-protein coupled RV 13 52482227 Drd1a opposite downstream 133665 s07 D1A receptor 89755C RV 13 56843379 Ubqln1 ubiquilin 1 same intragenic 1012 In1 Miscellaneous function 08 89755D death associated Non-receptor serine/threonine RV 13 59329938 Dapk1 same intragenic 69090 In1 02 protein kinase 1 protein kinase RV 89754A 13 72466712 2810465 RIKEN cDNA same intragenic 23953 In1 unclassified
28 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 07 F10Rik 2810465F10 gene coagulation factor II 89755B Receptor, G-protein coupled RV 13 91875486 F2r12 (thrombin) receptor- opposite upstream 11859 03 receptor like 2 mitogen activated 89754D RV 13 108200362 Map3k1 protein kinase kinase same upstream 1128 Kinase, Protein kinase 3 - 05 kinase 1 89792A potassium channel, RV 14 18536544 Kcnk5 opposite intragenic 7973 In1 Ion channel, Other ion channel 3 CIS 07 subfamily K, member 5 89754E D930025 hypothetical protein RV 14 28778762 opposite upstream 11871 unclassified s07 H04 D930025H04 Signaling molecule, 89754H ankyrin repeat domain RV 14 29912665 Ankrd28 same intragenic 49752 In1 Transmembrane receptor 06 28 regulatory/adaptor protein 89792D granule cell antiserum RV 14 35081867 Gcap14 same upstream 18425 unclassified 08 positive 14 dehydrogenase/reducta 89755C Oxidoreductase, Dehydrogenase, RV 14 49904878 Dhrs4 se (SDR family) opposite upstream 91052 04 Reductase member 4 89755H methionine sulfoxide RV 14 58969592 Msra same intragenic 14590 In5 Oxidoreductase, Reductase 1 - 10 reductase A 89791H Receptor, Oxidoreductase, RV 14 63982955 Lox12 lysyl oxidase-like 2 opposite intragenic 57557 In5 07 Oxidase 89755C Transcription factor , Nucleic acid RV 14 67677860 Rb1 retinoblastoma 1 same upstream 684 12 binding 89755A BC05232 cDNA sequence RV 15 27685388 opposite upstream 15327 unclassified 08 8 BC052328 89791F 3110006 RV 15 36903779 zinc finger protein 706 opposite downstream 96164 unclassified 07 P09Rik melanoma-derived 89792D RV 15 62626408 Mlze leucine zipper, extra- same downstream 165858 unclassified 2 - 07 nuclear factor
29 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene
ST3 beta-galactoside 89791G RV 15 67128256 St3gal1 alpha-2,3- same intragenic 9943 Ex6 Transferase, Glycosyltransferase 4 - 04 sialyltransferase 1 89791D Tbc1d22 TBC1 domain family, RV 15 86395301 same intragenic 129957 In7 unclassified 2 - 04 a member 22a 89792B Cytoskeletal protein, Actin family RV 15 99400526 Fmnl3 formin-like 3 same intragenic 27746 In1 3 CIS 01 cytoskeletal protein 89754F RV 15 102292356 Itgb7 integrin beta 7 opposite upstream 1109 Receptor, Cell adhesion molecule 07 89754G RV 16 16928792 Rtn4r reticulon 4 receptor opposite downstream 5343 Receptor 12 UDP-GlcNAc:betaGal 89754A beta-1,3-N- RV 16 18531775 B3gnt5 same intragenic 202 Ex1 Transferase, Glycosyltransferase 05 acetylglucosaminyltran sferase 5 89755F interferon (alpha and Cytokine receptor, Interferon RV 16 90528527 Ifnar2 opposite upstream 2111 1 - 11 beta) receptor 2 receptor 89754C interferon (alpha and Cytokine receptor, Interferon RV 16 90679548 Ifnar1 same downstream 15102 05 beta) receptor 1 receptor Transcription factor, Other 89791G runt related RV 16 91974100 Runx1 opposite upstream 115686 transcription factor, Nucleic acid 7 CIS 01 transcription factor 1 binding avian erythroblastosis 89754C Transcription factor , Nucleic acid RV 16 94848287 Erg virus E-26 (v-ets) same intragenic 89802 In2 2 - 08 binding oncogene related Down syndrome 89791B RV 16 95427835 Dscr2 critical region homolog same intragenic 473 In1 unclassified 03 2 (human) 89791A 5730437 RIKEN cDNA RV 17 5103292 opposite downstream 214958 unclassified 1 - 09 N04Rik 5730437N04 gene 89754C RV 17 24767940 Zbtb9 same upstream 303 unclassified 04
30 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene solute carrier family 37 89792H RV 17 29098106 Slc37a1 (glycerol-3-phosphate same upstream 5244 Transporter, Other transporter 02 transporter), member 1 Receptor, Signaling molecule, 89792B Notch gene homolog 3 Membrane-bound signaling RV 17 29915608 Notch3 same downstream 13218 07 (Drosophila) molecule, Defense/immunity protein complement Protease, Serine protease, 89754A RV 17 32581814 C2 component 2 (within same intragenic 3053 In3 Defense/immunity protein, 01 H-2S) Complement component 89792F suppressor of Ty 3 Transcription factor, Basal RV 17 42624522 Supt3h same downstream 155 1 - 05 homolog (S. cerevisiae) transcription factor 89791C elastin microfibril Extracellular matrix, Extracellular RV 17 69083907 Emilin2 opposite intragenic 234 Ex2 1 - 04 interfacer 2 matrix glycoprotein 89754A 1110006 RIKEN cDNA RV 18 35969126 same upstream 23969 unclassified 12 O17Rik 1110006O17 gene 89754F fem-1 homolog c RV 18 46746414 Fem1c opposite intragenic 1314 Ex2 unclassified 04 (C.elegans) 89792C casein kinase 1, gamma Non-receptor serine/threonine RV 18 54122341 Csnk1g3 same upstream 3285 1 - 11 3 protein kinase 89755F membrane-associated RV 18 57191667 March3 opposite upstream 48199 unclassified 01 ring finger (C3HC4) 3 89754G isochorismatase RV 18 58793038 Isoc1 same upstream 91007 unclassified 05 domain containing 1 eukaryotic translation 89791F Nucleic acid binding, Translation RV 19 8179771 Eef1g elongation factor 1 opposite downstream 5045 1 - 10 elongation factor gamma 89791G solute carrier family RV 19 10039872 Slc15a3 same intragenic 830 Ex1 Transporter, Other transporter 03 15, member 3 89792H Non-receptor tyrosine protein RV 19 28505911 Jak2 Janus kinase 2 same intragenic 856 In1 1 - 07 kinase 89755D transmembrane protein RV 19 31511701 Tmem23 same intragenic 61809 In2 unclassified 4 CIS 01 23 89754A transmembrane protein RV 19 31592608 Tmem23 same upstream 19098 unclassified 4 CIS 04 23
31 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89791G nebulin-related Actin family cytoskeletal protein, RV 19 55975059 Nrap opposite upstream 404 09 anchoring protein Non-motor actin binding protein Select calcium binding protein , 89754C dystrophin, muscular RV X 78247932 Dmd same intragenic 638372 In7 Actin family cytoskeletal protein, 11 dystrophy Non-motor actin binding protein 89791D kelch-like 15 RV X 88896176 Klhl15 opposite intragenic 513 In2 unclassified 05 (Drosophila) 89792E RV X 127656314 Nox1 NADPH oxidase 1 opposite intragenic 334 In1 Oxidoreductase, Oxidase s08 89791C AW5471 expressed sequence RV X 136019057 opposite upstream 102405 unclassified 02 86 AW547186 89791E RV SINE, B1, family B4 s04 89792E RV multiple hits s04 89754B RV LTR, ERVK 03 89754H RV simple repeat / SINE 02 89755A RV LTR, ERVK 01 89755A RV LINE 02 89755B RV multiple hits 02 89755B RV multiple hits 10 89755C RV SINE 09 89755D RV LTR, ERVL 11 89791B RV repeat 04 89791B RV LTR, MaLR, MLT1D 06 89791D RV SINE, Alu, B1_Mus1 02
32 Supplementary Table 3; Montini E. et al. 2006
Integratio Nearest ORI Molecular Function Vect SEQ- Position Distance to RTCGD RTCG Chr n position Gene Nearest Gene Name vector/ or NO. Relative to Gene TSS (bp) Hits D CIS (bp) Symbol gene 89791D RV repeat 03 89791D LTR, ERVK, BGLII- RV 09 int 89791F RV multiple hits 01 89792G RV multiple hits 11
The genomic part of each retrieved LAM-PCR amplicons obtained from LV (LV pre ) and RV-transduced (RV pre) pretransplant cells was mapped to the murine genome using the UCSC Santa Cruz BLAT alignment tools (www.genome.ucsc.edu), Mouse Assembly mm7. Seq-No., sequence identity; Chr, chromosome; Integration position (bp), position of vector integration into the chromosome; Nearest Gene Symbol and Nearest Gene Name, nearest gene obtained from RefSeq or Locus Link DBs at UCSC, integrations landing into repeats or low complexity regions with multiple positions in the genome are indicated; ORI vector/gene, orientation of vector LTRs with respect the direction of transcription of the nearest gene; Position Relative to Gene, integrations localized siding the 5’ or the 3’ of the nearest gene are classified as upstream or downstream respectively, integrations inside genes are intragenic; Distance to TSS (bp), was calculated by subtracting chromosomal coordinates of vector integration to the transcript start coordinates of the respective nearest gene (gene coordinates obtained from mm6 UCSC table annotations), for upstream integrations distance is the absolute number if the subtraction, for intragenic integrations the intron/exon number were vector integrated is also indicated; Molecular function, annotation of each gene was obtained using the Celera Discovery System (http://myscience.appliedbiosystems.com); RTCGD Hits, number of times the gene was retrieved in experiments reported by the retroviral tagged cancer gene database; RTCGD CIS, genes listed as common integration sites (CIS), (-) non-CIS genes listed in the database.
33