Supplementary Material
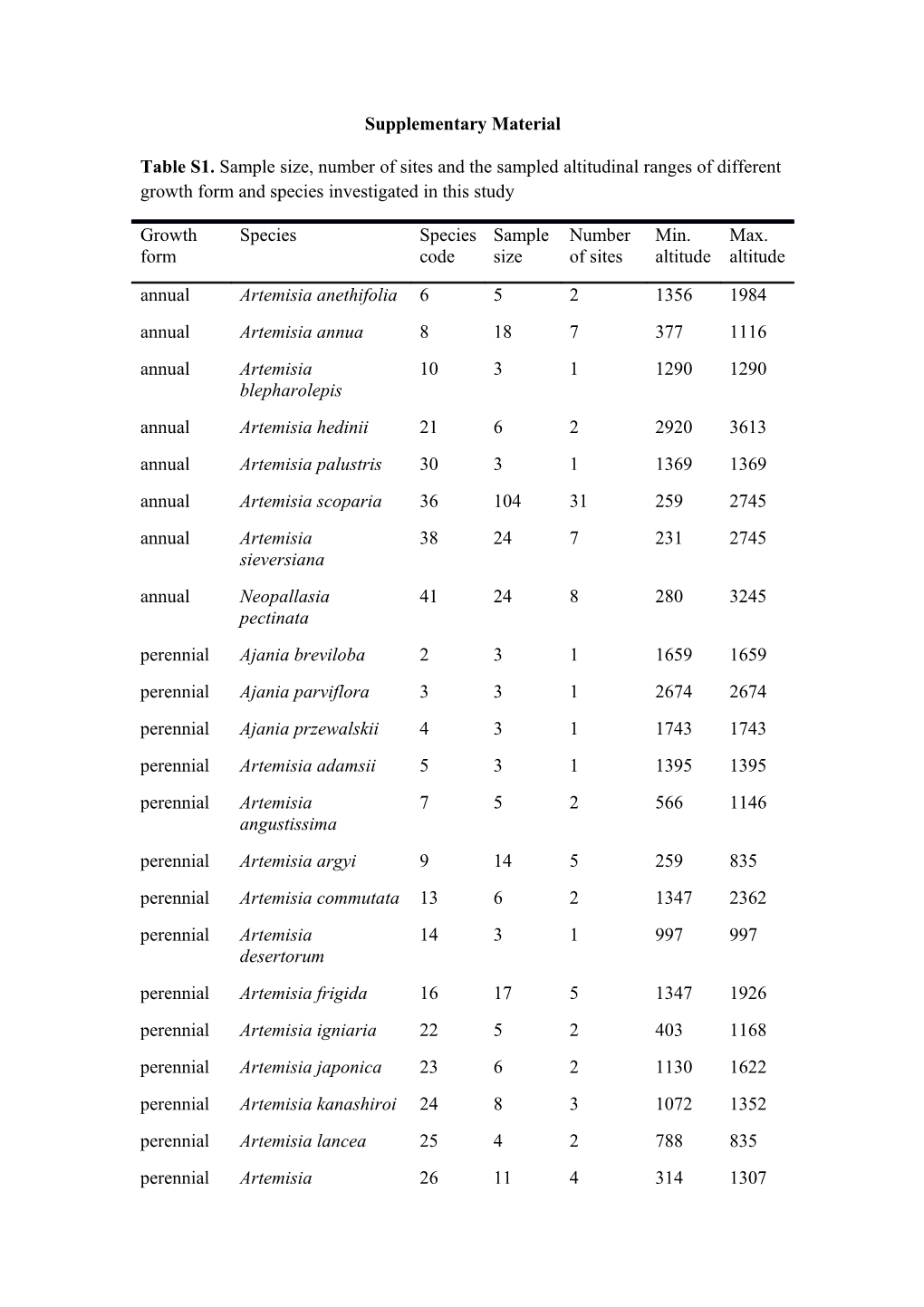
Table S1. Sample size, number of sites and the sampled altitudinal ranges of different growth form and species investigated in this study
Growth Species Species Sample Number Min. Max. form code size of sites altitude altitude annual Artemisia anethifolia 6 5 2 1356 1984 annual Artemisia annua 8 18 7 377 1116 annual Artemisia 10 3 1 1290 1290 blepharolepis annual Artemisia hedinii 21 6 2 2920 3613 annual Artemisia palustris 30 3 1 1369 1369 annual Artemisia scoparia 36 104 31 259 2745 annual Artemisia 38 24 7 231 2745 sieversiana annual Neopallasia 41 24 8 280 3245 pectinata perennial Ajania breviloba 2 3 1 1659 1659 perennial Ajania parviflora 3 3 1 2674 2674 perennial Ajania przewalskii 4 3 1 1743 1743 perennial Artemisia adamsii 5 3 1 1395 1395 perennial Artemisia 7 5 2 566 1146 angustissima perennial Artemisia argyi 9 14 5 259 835 perennial Artemisia commutata 13 6 2 1347 2362 perennial Artemisia 14 3 1 997 997 desertorum perennial Artemisia frigida 16 17 5 1347 1926 perennial Artemisia igniaria 22 5 2 403 1168 perennial Artemisia japonica 23 6 2 1130 1622 perennial Artemisia kanashiroi 24 8 3 1072 1352 perennial Artemisia lancea 25 4 2 788 835 perennial Artemisia 26 11 4 314 1307 lavandulifolia perennial Artemisia 27 9 3 1113 2674 leucophylla perennial Artemisia mongolica 28 48 15 231 3245 perennial Artemisia phaeolepis 31 2 1 3635 3635 perennial Artemisia pubescens 32 12 4 1369 1984 perennial Artemisia rubripes 34 62 19 259 2920 perennial Artemisia 37 3 1 314 314 selengensis perennial Artemisia subulata 39 3 1 1116 1116 perennial Seriphidium 45 5 2 2420 3245 nitrosum perennial Seriphidium terrae- 47 8 3 1743 3635 albae semi- Ajania achilloides 1 3 1 1926 1926 shrub semi- Artemisia 11 9 3 1926 3613 shrub brachyloba semi- Artemisia capillaris 12 21 6 403 2674 shrub semi- Artemisia dubia 15 2 1 1352 1352 shrub semi- Artemisia gansuensis 17 3 1 1732 1732 shrub semi- Artemisia giraldii 18 23 8 311 2420 shrub semi- Artemisia gmelinii 19 2 1 2674 2674 shrub semi- Artemisia 20 3 1 1088 1088 shrub halodendron semi- Artemisia ordosica 29 12 4 1307 1515 shrub semi- Artemisia 33 3 1 3613 3613 shrub roxburghiana semi- Artemisia sacrorum 35 48 16 377 2920 shrub semi- Hippolytia trifida 40 3 1 1732 1732 shrub semi- Seriphidium finitum 42 2 1 902 902 shrub semi- Seriphidium 43 3 1 2851 2851 shrub gracilescens semi- Seriphidium 44 3 1 1743 1743 shrub lehmannianum semi- Seriphidium 46 5 1 1587 1587 shrub santolinum Table S2. Effects of annual climate on non-structural carbohydrates (NSC) of Artemisia species and their close relatives across northern China
Variable Predictor Estimate t- value
REML MCMC
NSC Intercept 33.573(19.532) 32.862(-5.672,68.342) 1.719 AMT -18.438(11.125) -18.028(-38.318,4.015) -1.657 AP -11.783(7.616) -11.499(-26.852,2.652) -1.547 AMT*AP 6.771(4.336) 6.607(-1.352,15.471) 1.562 Sugar Intercept 38.820(34.917) 37.705(-30.312,101.812) 1.112 AMT -21.797(19.885) -21.160(-58.032,17.313) -1.096 AP -14.086(13.640) -13.648(-38.512,13.542) -1.033 AMT*AP 8.078(7.765) 7.828(-7.711,21.872) 1.040 Starch Intercept 31.284(18.063) 32.292(-4.219, 67.801) 1.732 AMT -17.189(10.291) -17.760(-38.164,2.966) -1.670 AP -11.021(7.019) -11.422(-24.796,3.246) -1.570 AMT*AP 6.328(3.997) 6.556(-1.912,14.112) 1.583
Restricted maximum likelihood (REML, estimate with standard error in parentheses) and Markov chain Monte Carlo (MCMC, estimate with HPD 95% intervals in parentheses) parameter estimates and their statistical significance for the fixed effects. AMT, annual mean temperature; AP, annual precipitation. R script for statistical analyses in this study
#### reading data#### dat=read.csv(###data file path###,header=T)
##constructing a data frame that contains data used in this study## dat1=cbind.data.frame(dat$location,dat$life.form,dat$species,dat$C.mg.g,dat$N.mg.g ,dat$sugar.mg.g,dat$starch.mg.g,dat$protein.mg.g,dat[,7:9],dat[,44:46],dat[,48:66]) colnames(dat1) [1:8]=c("location","life.form","species","C.mg.g","N.mg.g","sugar.mg.g","starch.mg. g","protein.mg.g")
##calculating altitudinal ranges of each species### tab=c() spe=levels(dat1$species) for (i in 1:length(spe)){ lif=unique(dat1$life.form[dat1$species==spe[i]]) tab=rbind(tab,c(paste(lif),paste(spe[i]),range(dat1$altit[dat1$species==spe[i]])))
}
##calculating site number of each species### tab=c() spe=levels(dat1$species) for (i in 1:length(spe)){ dat2=subset(dat1,species==spe[i]) dat2$location=factor(dat2$location) tab=rbind(tab,paste(spe[i]),nlevels(unique(dat2$location)))
}
##calculating sample size of each species### tab=c() spe=levels(dat1$species) for (i in 1:length(spe)){ dat2=subset(dat1,species==spe[i]) dat2$location=factor(dat2$location) tab=rbind(tab,paste(spe[i]),nrow(dat2))
}
####calculating C constituents####
##Hoch et al. (2002): 0.41 g C per g sugar (mean of glucose, fructose and sucrose), 0.444 g C per g starch### dat1=na.exclude(dat1) dat1$nsc=0.41*dat1$sugar.mg.g+0.444*dat1$starch.mg.g dat1$sc=dat1$C.mg.g-dat1$nsc dat1$sug.c=0.41*dat1$sugar.mg.g dat1$star.c=0.444*dat1$starch.mg.g dat1$ratio=dat1$sugar.mg.g/dat1$starch.mg.g dat1$total=dat1$sugar.mg.g+dat1$starch.mg.g
##descriptive analyses### mean(dat1$total) mean(dat1$sugar) mean(dat1$starch) mean(dat1$ratio) sd(dat1$total) sd(dat1$sugar) sd(dat1$starch) sd(dat1$ratio)
##log transformation### dat1$C=log10(dat1$C.mg.g) dat1$sugar=log10(dat1$sugar.mg.g) dat1$starch=log10(dat1$starch.mg.g) dat1$ratio=log10(dat1$ratio) dat1$total=log10(dat1$total) dat1$nsc=log10(dat1$nsc) dat1$sc=log10(dat1$sc) dat1$sug.c=log10(dat1$sug.c) dat1$star.c=log10(dat1$star.c)
###hierarchical models### library(languageR) library(lme4) library(MCMCglmm)
##geographical varible## dat1$longit=log10(dat1$longit) dat1$latit=log10(dat1$latit) dat1$altit=log10(dat1$altit) carb=c(colnames(dat1[,34:42])) for (i in 1:length(carb)){ form=formula(paste(carb[i],"~longit+latit+altit+(1|location)",sep="")) dat.lmer=lmer(form,data=dat1) print(paste("##",carb[i],sep="")) print(summary(dat.lmer)) form=formula(paste(carb[i],"~longit+latit+altit",sep="")) dat.mc=MCMCglmm(form,, random=~location,nitt=10000,data=dat1, verbose=FALSE) print(summary(dat.mc))
}
##climate effects ### dat1$bio1=log10(dat1$bio1+50) dat1$bio12=log10(dat1$bio12) dat1$bio10=log10(dat1$bio10) dat1$bio18=log10(dat1$bio18)
#annual climate# for (i in 1:length(carb)){ form=formula(paste(carb[i],"~bio1*bio12+(1|location)",sep="")) dat.lmer=lmer(form,data=dat1) print(paste("##",carb[i],sep="")) print(summary(dat.lmer)) form=formula(paste(carb[i],"~bio1*bio12",sep="")) dat.mc=MCMCglmm(form,, random=~location,nitt=10000,data=dat1, verbose=FALSE) print(summary(dat.mc))
}
#warmest quarter climate# for (i in 1:length(carb)){ form=formula(paste(carb[i],"~bio10*bio18+(1|location)",sep="")) dat.lmer=lmer(form,data=dat1) print(paste("##",carb[i],sep="")) print(summary(dat.lmer)) form=formula(paste(carb[i],"~bio10*bio18",sep="")) dat.mc=MCMCglmm(form,, random=~location,nitt=10000,data=dat1, verbose=FALSE) print(summary(dat.mc))
}
####nested ANOVA### dat1$species=factor(dat1$species) dat1$life.form=factor(dat1$life.form) dat1$location=factor(dat1$location)
library(nlme) library(boot) library(ape)
##nested variance partition### for (i in 1:length(carb)){ form=formula(paste(carb[i],"~1",sep="")) dat.varcomp=varcomp(lme(form,random=~1|location/life.form/species,data=dat1),1) print(paste("####",carb[i],sep="")) print(dat.varcomp)
##boxplot to show relationships between carbon in sugar and starch## lf=levels(dat1$life.form) col1=c("blue","yellow","green") col2=c("light blue","light yellow","light green") dval=c(0,12,24) aval=c(30,45,60) boxplot(dat1$sug.c~dat1$species,ylim=c(-1,2),xlab="species",ylab="C concentration (log scale)",cex.lab=1.3,cex.axis=1.2,type="n") for (i in 1:length(lf)){ dat2=subset(dat1,life.form==lf[i]) outstat<-boxplot(dat2$sug.c~dat2$species,col=col1[i],labs=F,axes=F,add=T) outstat<-boxplot(dat2$star.c~dat2$species,col=col2[i],labs=F,axes=F,add=T) ngroups <- length(levels(dat2$species)) rect((1:ngroups)-.4, outstat$stats[2,], (1:ngroups)+.4, outstat$stats[4,],
density=dval[i], angle=aval[i])
}
legend("bottomright", paste(lf),pch=15,col=col1,title="Sugar") legend("topright", paste(lf),pt.bg=col2,angle=aval,density=dval,title="Starch")