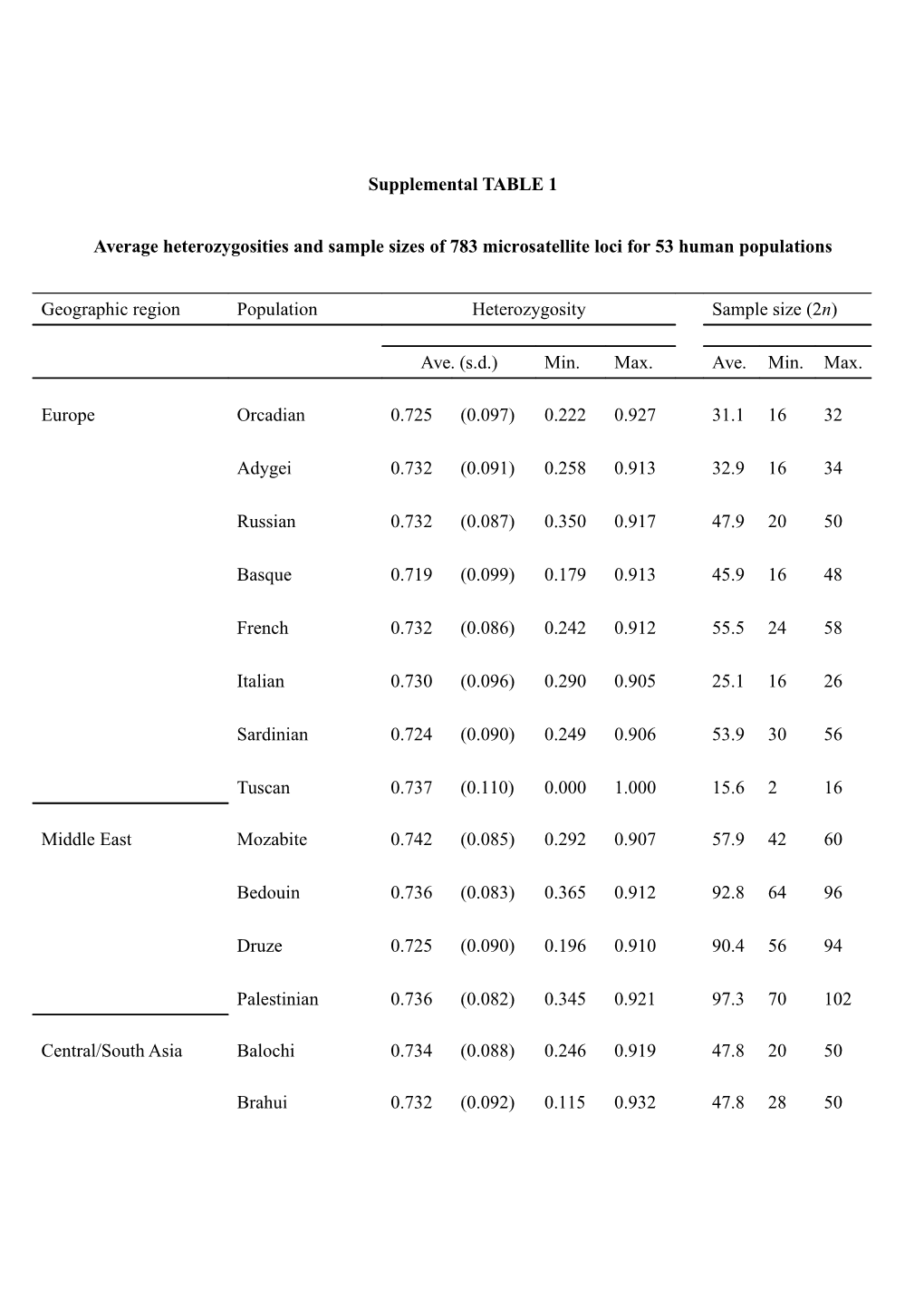
Supplemental TABLE 1
Average heterozygosities and sample sizes of 783 microsatellite loci for 53 human populations
Geographic region Population Heterozygosity Sample size (2n)
Ave. (s.d.) Min. Max. Ave. Min. Max.
Europe Orcadian 0.725 (0.097) 0.222 0.927 31.1 16 32
Adygei 0.732 (0.091) 0.258 0.913 32.9 16 34
Russian 0.732 (0.087) 0.350 0.917 47.9 20 50
Basque 0.719 (0.099) 0.179 0.913 45.9 16 48
French 0.732 (0.086) 0.242 0.912 55.5 24 58
Italian 0.730 (0.096) 0.290 0.905 25.1 16 26
Sardinian 0.724 (0.090) 0.249 0.906 53.9 30 56
Tuscan 0.737 (0.110) 0.000 1.000 15.6 2 16
Middle East Mozabite 0.742 (0.085) 0.292 0.907 57.9 42 60
Bedouin 0.736 (0.083) 0.365 0.912 92.8 64 96
Druze 0.725 (0.090) 0.196 0.910 90.4 56 94
Palestinian 0.736 (0.082) 0.345 0.921 97.3 70 102
Central/South Asia Balochi 0.734 (0.088) 0.246 0.919 47.8 20 50
Brahui 0.732 (0.092) 0.115 0.932 47.8 28 50 Burusho 0.730 (0.090) 0.246 0.919 48.8 32 50
Hazara 0.728 (0.096) 0.232 0.930 45.7 24 48
Kalash 0.699 (0.113) 0.000 0.908 48.3 26 50
Makrani 0.740 (0.089) 0.366 0.914 47.8 22 50
Pathan 0.735 (0.092) 0.198 0.932 46.2 28 48
Sindhi 0.735 (0.090) 0.115 0.916 48.4 34 50
Uygur 0.731 (0.113) 0.000 0.954 19.1 4 20
Oceania Melanesian 0.660 (0.146) 0.056 0.934 36.3 14 38
Papuan 0.674 (0.144) 0.000 0.923 32.7 10 34
America Colombian 0.604 (0.171) 0.000 0.892 25.1 16 26
Karitiana 0.552 (0.182) 0.000 0.858 46.6 26 48
Maya 0.670 (0.132) 0.000 0.911 48.3 32 50
Pima 0.598 (0.169) 0.000 0.857 48.6 32 50
Africa Bantu Kenya 0.759 (0.099) 0.301 0.957 23.1 24 16
Bantu South East 0.770 (0.124) 0.200 1.000 9.7 4 10
Bantu South West 0.761 (0.172) 0.000 1.000 5.8 2 6
Mandenka 0.756 (0.090) 0.159 0.946 46.3 48 32
Yoruba 0.760 (0.089) 0.222 0.940 48.4 30 50
Biaka Pygmy 0.759 (0.092) 0.097 0.938 62.2 18 64 Mbuti Pygmy 0.750 (0.105) 0.091 0.949 28.9 20 30
San 0.744 (0.129) 0.000 1.000 13.6 6 14
East Asia Han 0.705 (0.116) 0.058 0.924 65.8 40 68
Han Northern 0.709 (0.130) 0.000 0.963 19.3 6 20 China
Dai 0.699 (0.140) 0.000 0.935 19.1 4 20
Daur 0.708 (0.128) 0.000 0.942 19.2 6 20
Hezhen 0.705 (0.129) 0.000 0.945 17.1 2 18
Lahu 0.681 (0.143) 0.000 0.933 19.1 6 20
Miao 0.698 (0.138) 0.000 0.935 19.4 10 20
Oroqen 0.693 (0.135) 0.000 0.933 19.2 6 20
She 0.691 (0.132) 0.000 0.926 19.4 8 20
Tujia 0.699 (0.137) 0.000 0.912 19.1 6 20
Tu 0.706 (0.128) 0.000 0.926 19.4 10 20
Xibo 0.708 (0.132) 0.000 0.974 17.5 8 18
Mongola 0.711 (0.129) 0.100 0.947 19.2 4 20
Naxi 0.694 (0.132) 0.000 0.915 19.3 10 20
Cambodian 0.708 (0.131) 0.100 0.944 21.0 8 22
Japanese 0.701 (0.123) 0.000 0.929 56.0 36 58 Yakut 0.701 (0.107) 0.115 0.915 48.4 34 50
n is the number of individuals examined excluding missing data.
r Standard deviation of heterozygosity is computed by square root of V(h) = (h - H)2 /(r - 1) е j=1 j where h is a heterozygosty of a locus and r is the number of loci.