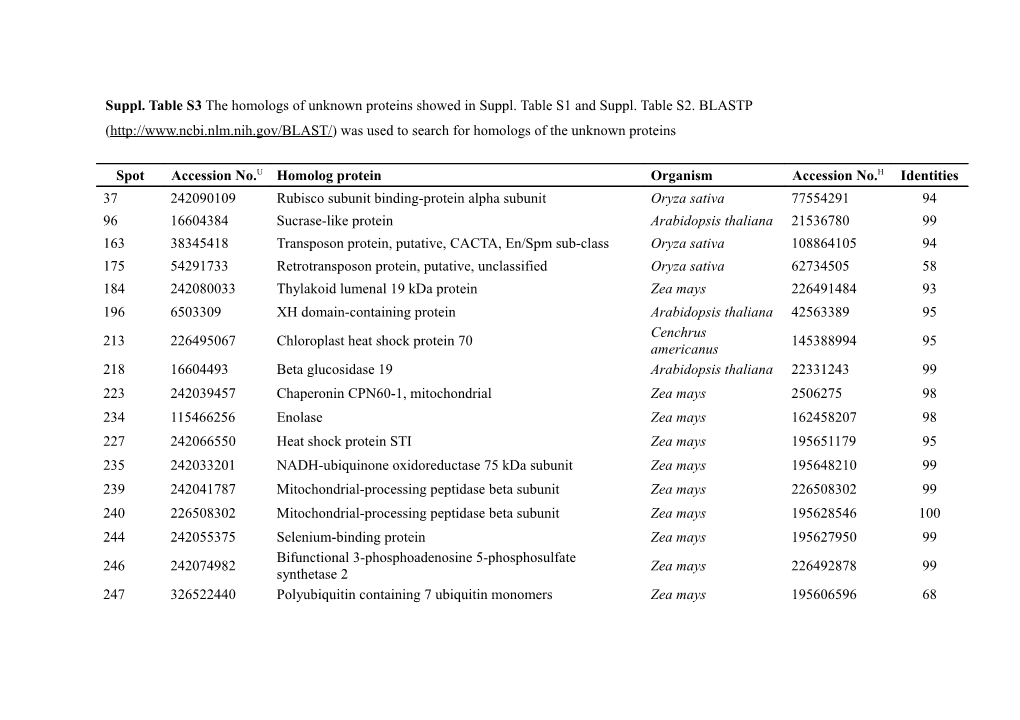
Suppl. Table S3 The homologs of unknown proteins showed in Suppl. Table S1 and Suppl. Table S2. BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/) was used to search for homologs of the unknown proteins
Spot Accession No.U Homolog protein Organism Accession No.H Identities 37 242090109 Rubisco subunit binding-protein alpha subunit Oryza sativa 77554291 94 96 16604384 Sucrase-like protein Arabidopsis thaliana 21536780 99 163 38345418 Transposon protein, putative, CACTA, En/Spm sub-class Oryza sativa 108864105 94 175 54291733 Retrotransposon protein, putative, unclassified Oryza sativa 62734505 58 184 242080033 Thylakoid lumenal 19 kDa protein Zea mays 226491484 93 196 6503309 XH domain-containing protein Arabidopsis thaliana 42563389 95 Cenchrus 213 226495067 Chloroplast heat shock protein 70 145388994 95 americanus 218 16604493 Beta glucosidase 19 Arabidopsis thaliana 22331243 99 223 242039457 Chaperonin CPN60-1, mitochondrial Zea mays 2506275 98 234 115466256 Enolase Zea mays 162458207 98 227 242066550 Heat shock protein STI Zea mays 195651179 95 235 242033201 NADH-ubiquinone oxidoreductase 75 kDa subunit Zea mays 195648210 99 239 242041787 Mitochondrial-processing peptidase beta subunit Zea mays 226508302 99 240 226508302 Mitochondrial-processing peptidase beta subunit Zea mays 195628546 100 244 242055375 Selenium-binding protein Zea mays 195627950 99 Bifunctional 3-phosphoadenosine 5-phosphosulfate 246 242074982 Zea mays 226492878 99 synthetase 2 247 326522440 Polyubiquitin containing 7 ubiquitin monomers Zea mays 195606596 68 249 224030739 Aldo/keto reductase family-like protein Oryza sativa 38175438 83 250 242039369 Malate dehydrogenase, cytoplasmic Zea mays 162464321 99 255 326512270 Heat shock factor protein 1 Zea mays ACG48787.1 60 262 242087799 50S ribosomal protein L1 Zea mays 226496743 94 265, 266 242084916 Magnesium-chelatase subunit chlI Zea mays ACG34535.1 97 269 242059967 Ferredoxin Zea mays 162458489 92 280 194689474 Aldo-keto reductase/ oxidoreductase Zea mays ACG34358.1 98 293 242073212 22.0 kDa class IV heat shock protein Zea mays 226509936 87 294 242041463 17.4 kDa class I heat shock protein 3 Zea mays 195626536 97 295 242052185 16.9 kDa class I heat shock protein 1 Zea mays 226505618 91 296 242052187 16.9 kDa class I heat shock protein 1 Zea mays 226505618 83 301 242072105 Rubisco activase 1, chloroplast precursor, putative Ricinus communis 255584538 75 316 38347311 Sedoheptulose-1,7-bisphosphatase precursor Oryza sativa 27804768 99 111 20821380P Putative bZIP transcription factor superfamily protein Zea mays 413944277 100 115 1956510P Cyclic nucleotide-gated ion channel 2 Arabidopsis thaliana 15242291 100 117 1956510P GAGA-binding protein Zea mays 226505882 82 155 19648253P BTB/POZ/Kelch-associated protein Arabidopsis thaliana 42566222 100 197 24139000P ethylene-binding protein-like Oryza sativa 51536200 100 217 20821380P putative bZIP transcription factor superfamily protein Zea mays 413944277 100 47 24139000P ethylene-binding protein-like Oryza sativa 51536200 100 Accession U, the accession number of the unknown proteins in Suppl. Table S1; Accession H, the accession number of the homologs identified by NCBI BLAST; P, The accession number in Phytozome database