Supplementary material
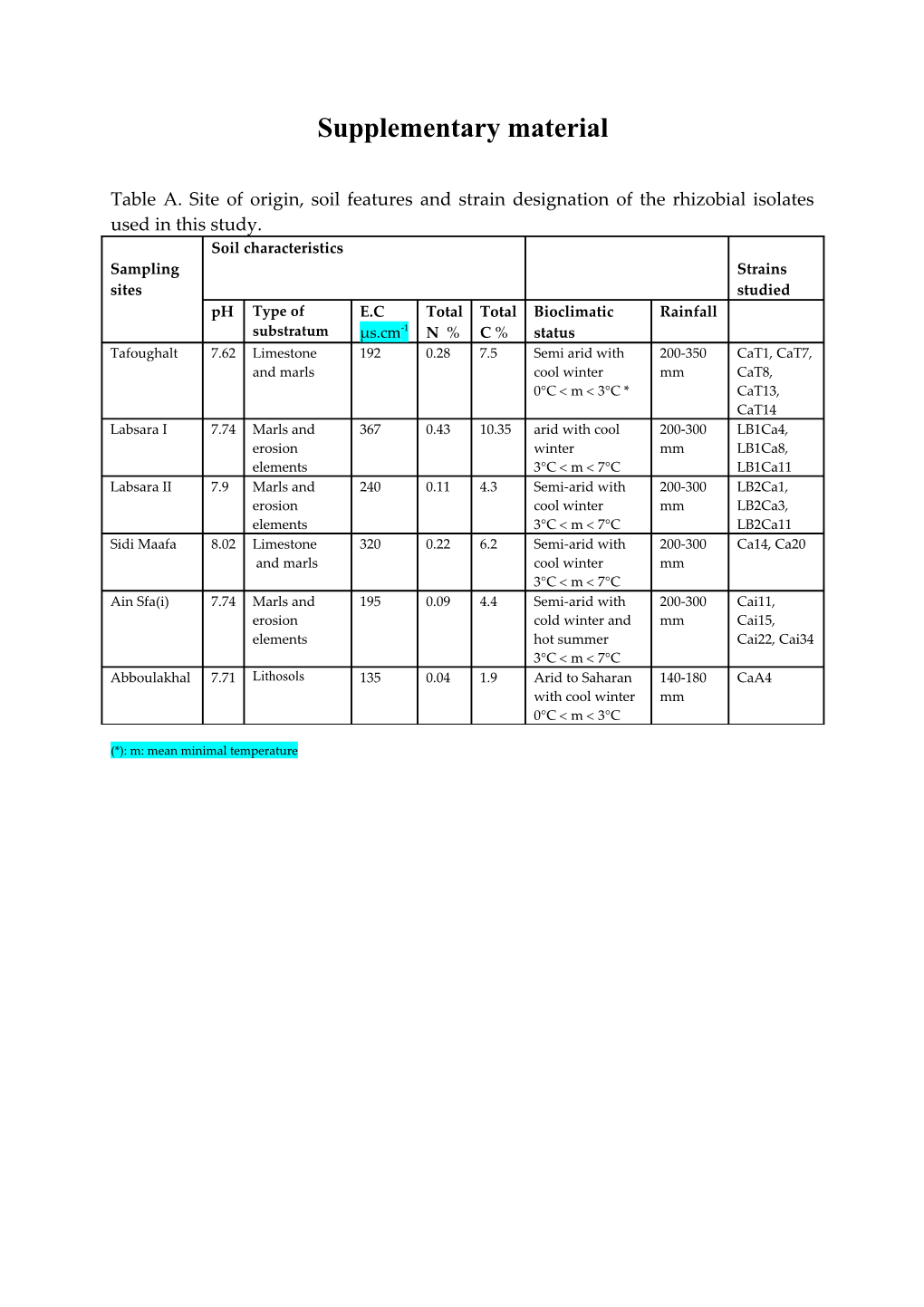
Table A. Site of origin, soil features and strain designation of the rhizobial isolates used in this study. Soil characteristics Sampling Strains sites studied pH Type of E.C Total Total Bioclimatic Rainfall substratum µs.cm-1 N % C % status Tafoughalt 7.62 Limestone 192 0.28 7.5 Semi arid with 200-350 CaT1, CaT7, and marls cool winter mm CaT8, 0°C m 3°C * CaT13, CaT14 Labsara I 7.74 Marls and 367 0.43 10.35 arid with cool 200-300 LB1Ca4, erosion winter mm LB1Ca8, elements 3°C m 7°C LB1Ca11 Labsara II 7.9 Marls and 240 0.11 4.3 Semi-arid with 200-300 LB2Ca1, erosion cool winter mm LB2Ca3, elements 3°C m 7°C LB2Ca11 Sidi Maafa 8.02 Limestone 320 0.22 6.2 Semi-arid with 200-300 Ca14, Ca20 and marls cool winter mm 3°C m 7°C Ain Sfa(i) 7.74 Marls and 195 0.09 4.4 Semi-arid with 200-300 Cai11, erosion cold winter and mm Cai15, elements hot summer Cai22, Cai34 3°C m 7°C Abboulakhal 7.71 Lithosols 135 0.04 1.9 Arid to Saharan 140-180 CaA4 with cool winter mm 0°C m 3°C
(*): m: mean minimal temperature Table B. Results of the biochemical and physiological tests used for the clustering analysis
Characteristic (a)A ((b)n =1) B (n = 9) C (n = 5) D (n = 3) Ribotype II Ribotypes I, Ribotypes Ribotypes I II, IV and V III, IV and V and IV NaCl tolerance 0% +(c) + + + 0.5% + + + + 1% + + 2 + 2% - (d) + 2 + 3% - + 1 + 4% - 2 - 1 5% - 1 - - 6% - - - - Growth at 25°C + + + + 30°C + + + + 35°C + + + + 40°C + 4 + + 43°C - 4 4 + 45°C - 1 2 1 Growth at pH 5.0 - 3 - - 5.5 - 8 - + 6.0 + + + + 7.0 + + + + 8.0 + + + + 9.0 + + + + 9.5 + 5 - 1 10.0 - 5 - 1 Growth at PEG 6000 0% + + + + 5% + + + + 10% + + + + 15% + + + + 20% + + + + 25% - + 3 + Heavy metals resistance (µg ml-1)
CuSO4, 5H2O (500) - + 4 2
HgCl2 (5) - 8 4 +
AlCl3(100) - + 3 1
AlCl3 (450) - 1 - -
CdSO4 (50) + + 4 +
ZnSO4 (250) + + 3 +
MgSO4 (1000) - 8 4 +
MgCl2 (1000) + + + +
MnSO4 (500) - + - +
MnCl2, 4H2O (500) + + + + Lead–acetate (1000) + + + +
BaCl2, 2H2O (1000) + + 4 +
CoSO4 (150) - 6 2 1
NiCl2, 6H2O (250) - + 3 2 Intrinsic antibiotic resistance (µg ml-1) Nalidixic acid (20) + + - + Rifampicin (20) + 4 - + Carbamicillin (20) + + - + Chloramphenicol (30) - 8 1 2 Gentamycin (10) + 1 - - Geniticin (20) + 8 - - Neomycin (20) + 1 - - Spectinomycin (20) + + - + Tetracyclin (5) + 8 - 2 Ampicillin (20) + + 2 + Carbohydrate utilization (1% w⁄ v) L-Rhamnose + + 2 2 D-Galactase + + 3 1 Meso-inositol + + 4 1 D-Mannose + + 3 - D-Fructose + + 1 1 Calcium citrate - 7 - - L-Arabinose + + 1 - D-Maltose + + 2 - Starch - 2 1 - Lactose - + 2 - Sucrose + + 3 - Raffinose + + 4 + Dextrins - 8 - 1 Trehalose + + - - D-Glucose + + - - Ribose + + 3 2 Sorbitol + + - - Salicin - 8 1 - Biochemical reactions Nitrate reduction + 4 2 2 Hydrolysis of urea - + + + Melanin production - 7 2 - Gelatinase activity + 3 2 2 Catalase activity + + + + Oxydase activity + + + +
(a) See Fig. 2 for clusters details. (b) (n) total number of strains tested per cluster. Numbers in the table are the number of strains able to grow at the biochemical test. (c) (-): all strains are sensitive. (d) (+): all strains are tolerant. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
Figure A. Examples of electrophoretic patterns generated by rep-PCR using REP primer: REP-PCR patterns obtained with the same isolates. Lanes: (1), molecular weight marker (200 bp ladder); (2) Ca14; (3) Ca20; (4) CaA4; (5) Cai11; (6) Cai15; (7) Cai22; (8) Cai34; (9) CaT1; (10) CaT7; (11) molecular weight marker (200 Pb ladder); (12) CaT8; (13) CaT13; (14) CaT14; (15) LB2Ca1; (16) LB2Ca3; (17) LB2Ca11; (18) LB1Ca4; (19) LB1Ca8.