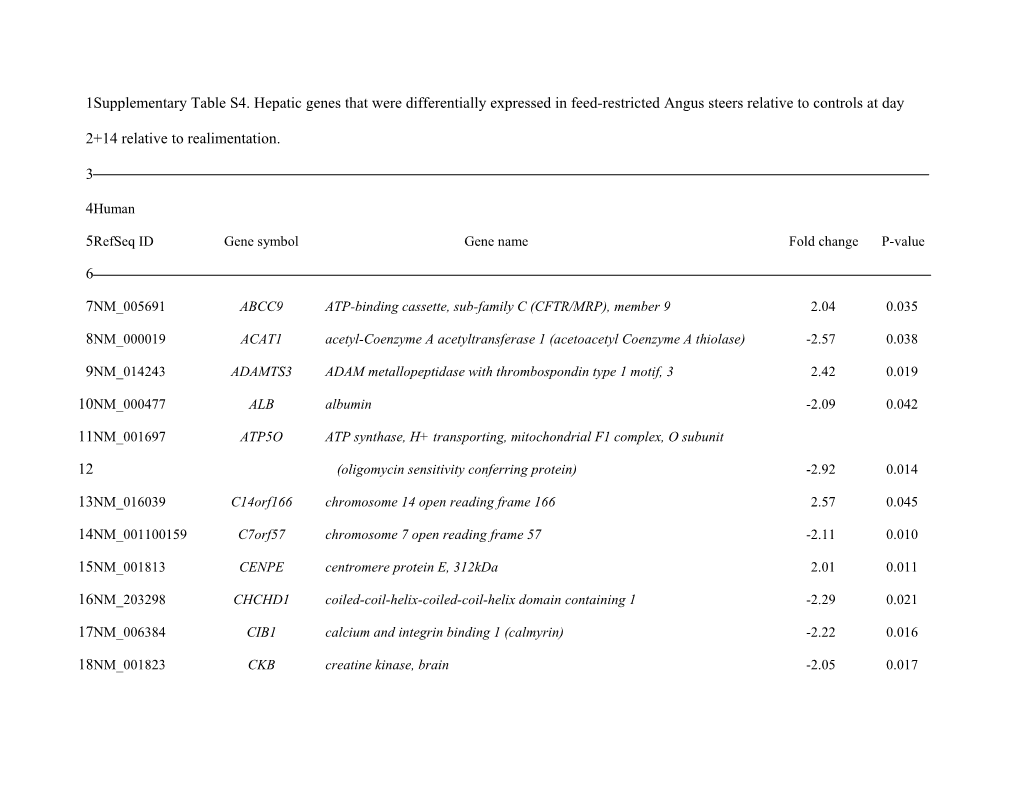
1Supplementary Table S4. Hepatic genes that were differentially expressed in feed-restricted Angus steers relative to controls at day
2+14 relative to realimentation.
3
4Human
5RefSeq ID Gene symbol Gene name Fold change P-value
6
7NM_005691 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 2.04 0.035
8NM_000019 ACAT1 acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) -2.57 0.038
9NM_014243 ADAMTS3 ADAM metallopeptidase with thrombospondin type 1 motif, 3 2.42 0.019
10NM_000477 ALB albumin -2.09 0.042
11NM_001697 ATP5O ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit
12 (oligomycin sensitivity conferring protein) -2.92 0.014
13NM_016039 C14orf166 chromosome 14 open reading frame 166 2.57 0.045
14NM_001100159 C7orf57 chromosome 7 open reading frame 57 -2.11 0.010
15NM_001813 CENPE centromere protein E, 312kDa 2.01 0.011
16NM_203298 CHCHD1 coiled-coil-helix-coiled-coil-helix domain containing 1 -2.29 0.021
17NM_006384 CIB1 calcium and integrin binding 1 (calmyrin) -2.22 0.016
18NM_001823 CKB creatine kinase, brain -2.05 0.017 1NM_014358 CLEC4E C-type lectin domain family 4, member E 2.15 0.021
2NM_138809 CMBL carboxymethylenebutenolidase homolog (Pseudomonas) -2.14 0.022
3NM_003418 CNBP CCHC-type zinc finger, nucleic acid binding protein -2.90 0.035
4NM_033381 COL4A5 collagen, type IV, alpha 5 (Alport syndrome) -2.08 0.038
5NM_014992 DAAM1 dishevelled associated activator of morphogenesis 1 -2.08 0.004
6NM_016306 DNAJB11 DnaJ (Hsp40) homolog, subfamily B, member 11 2.15 0.037
7NM_139072 DNER delta/notch-like EGF repeat containing -2.16 0.039
8NM_001011546 DSTN destrin (actin depolymerizing factor) -2.12 0.039
9NM_018098 ECT2 epithelial cell transforming sequence 2 oncogene 2.21 0.008
10NM_001970 EIF5A eukaryotic translation initiation factor 5A -2.25 0.027
11NM_000124 ERCC6 excision repair cross-complementing rodent repair deficiency, complementation
12 group 6 2.07 0.040
13NM_017669 ERCC6L excision repair cross-complementing rodent repair deficiency
14 complementation group 6-like 2.11 0.012
15NM_001031711 ERGIC endoplasmic reticulum-golgi intermediate compartment 1 -2.01 0.005
16NM_005243 EWSR1 Ewing sarcoma breakpoint region 1 2.01 0.031
17NM_016042 EXOSC3 exosome component 3 -2.37 0.003
18NM_015292 FAM62A family with sequence similarity 62 (C2 domain containing), member A 2.08 0.030 1NM_057092 FKBP2 FK506 binding protein 2, 13kDa -2.73 0.004
2NM_001002294 FMO3 flavin containing monooxygenase 3 -2.54 0.037
3NM_001039547 GK5 glycerol kinase 5 (putative) -2.75 0.003
4NM_025196 GRPEL1 GrpE-like 1, mitochondrial (E. coli) -2.00 0.044
5NM_147148 GSTM4 glutathione S-transferase M4 2.09 0.028
6NM_144594 GTSF1 gametocyte specific factor 1 2.25 0.005
7NM_016315 GULP1 GULP, engulfment adaptor PTB domain containing 1 2.26 0.003
8NM_014362 HIBCH 3-hydroxyisobutyryl-Coenzyme A hydrolase 2.13 0.018
9NM_020056 BOLA-DQA2 major histocompatibility complex, class II, DQ alpha 2 2.39 0.023
10NM_002130 HMGCS1 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) -2.75 0.026
11NM_005347 HSPA5 heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) -3.94 0.028
12NM_003856 IL1RL1 interleukin 1 receptor-like 1 2.14 0.037
13NM_144981 IMMP1L IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) 2.24 0.017
14XR_017711 LOC205251 LOC205251 -2.53 0.018
15XM_001714544 LOC338756 hypothetical LOC338 -2.12 0.021
16NM_014060 MCTS1 malignant T cell amplified sequence 1 -2.03 0.041
17NM_014168 METTL5 methyltransferase like 5 -2.11 0.018
18NM_000179 MSH6 mutS homolog 6 (E. coli) -2.12 0.038 1NM_001040113 MYH11 myosin, heavy chain 11, smooth muscle -2.12 0.029
2NM_018728 MYO5C myosin VC -2.17 0.020
3NM_005746 NAMPT nicotinamide phosphoribosyltransferase -2.24 0.026
4NM_005385 NKTR natural killer-tumor recognition sequence -2.06 0.008
5NM_003489 NRIP1 nuclear receptor interacting protein 1 -2.17 0.009
6NM_001079524 PAICS phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole
7 succinocarboxamide synthetase -2.27 0.020
8NM_006195 PBX3 pre-B-cell leukemia homeobox 3 2.16 0.024
9NM_031989 PCBP2 poly(rC) binding protein 2 -2.01 0.032
10NM_000285 PEPD peptidase D -2.01 0.048
11NM_002624 PFDN5 prefoldin subunit 5 -2.22 0.002
12NM_002625 PFKFB1 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 -2.47 0.029
13NM_016621 PHF21A PHD finger protein 21A -2.06 0.018
14NM_058179 PSAT1 phosphoserine aminotransferase 1 -2.19 0.034
15NM_001664 RHOA ras homolog gene family, member A -3.26 0.026
16NM_144563 RPIA ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) -2.00 0.029
17NM_016578 RSF1 remodeling and spacing factor 1 -2.02 0.001
18NM_153828 RTN4 reticulon 4 -2.27 0.016 1NM_198597 SEC24C SEC24 related gene family, member C (S. cerevisiae) -2.50 0.013
2NM_003016 SFRS2 splicing factor, arginine/serine-rich 2 -2.75 0.018
3NM_014031 SLC27A6 solute carrier family 27 (fatty acid transporter), member 6 -2.75 0.026
4NM_015191 SNF1LK2 SNF1-like kinase 2 -2.04 0.012
5NM_003089 SNRP70 small nuclear ribonucleoprotein 70kDa polypeptide (RNP antigen) -2.60 0.047
6NM_199436 SPAST spastin -2.03 0.014
7NM_152996 ST6GALNAC3 ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-
8 acetylgalactosaminide alpha-2,6-sialyltransferase 3 2.08 0.025
9NM_001039877 STRN4 striatin, calmodulin binding protein 4 -2.14 0.030
10NM_001008897 TCP1 t-complex 1 -2.28 0.010
11NM_018710 TMEM55A transmembrane protein 55A 2.21 0.015
12NM_003291 TPP2 tripeptidyl peptidase II -2.35 0.035
13NM_003292 TPR translocated promoter region (to activated MET oncogene) -2.43 0.017
14NM_000370 TTPA tocopherol (alpha) transfer protein (ataxia (Friedreich-like) with vitamin E
15 deficiency) -2.17 0.018
16