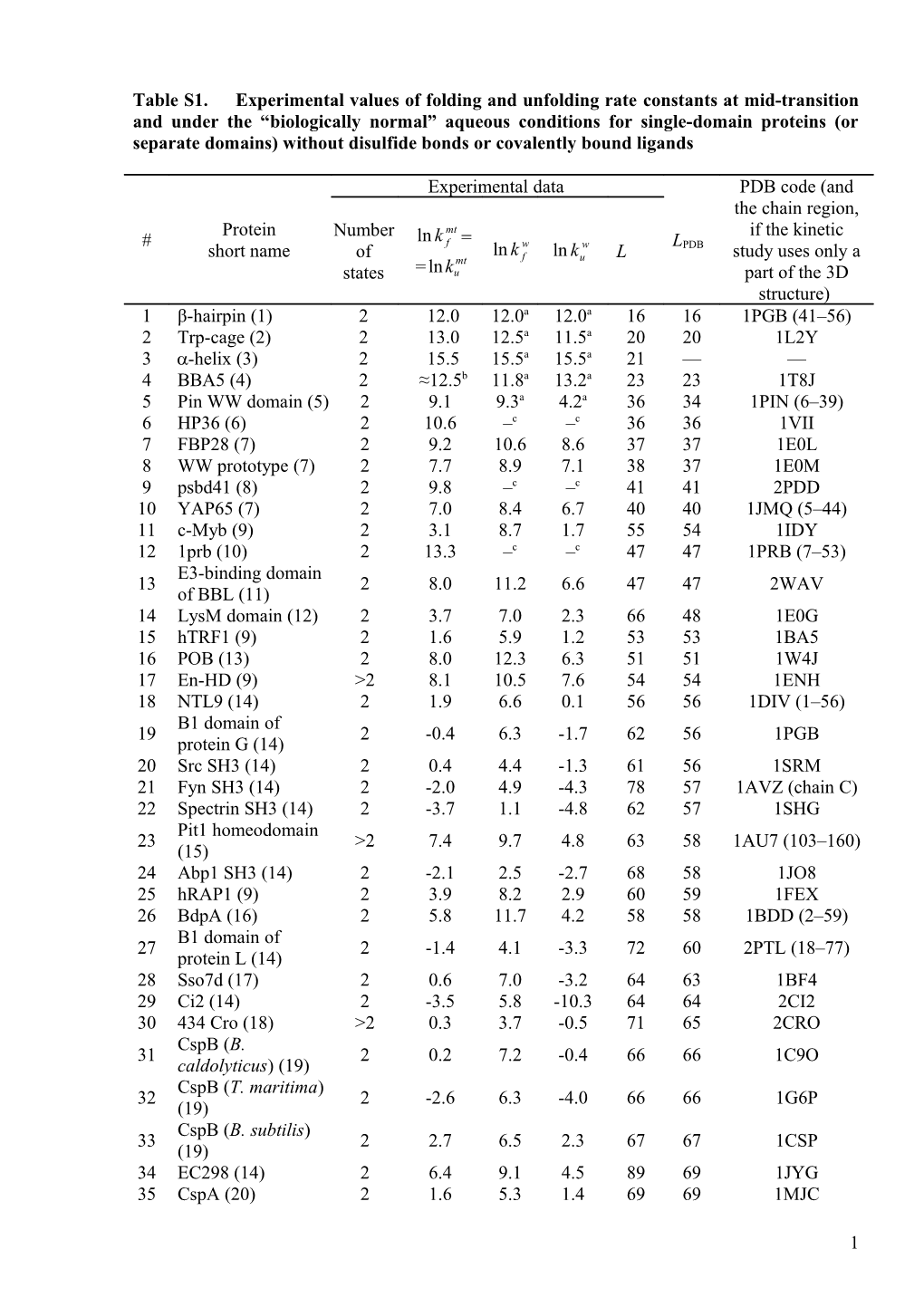
Table S1. Experimental values of folding and unfolding rate constants at mid-transition and under the “biologically normal” aqueous conditions for single-domain proteins (or separate domains) without disulfide bonds or covalently bound ligands
Experimental data PDB code (and the chain region, Protein Number mt if the kinetic # ln k f = w w L short name ln k ln k PDB study uses only a of mt f u L states =lnku part of the 3D structure) 1 β-hairpin (1) 2 12.0 12.0a 12.0a 16 16 1PGB (41–56) 2 Trp-cage (2) 2 13.0 12.5a 11.5a 20 20 1L2Y 3 -helix (3) 2 15.5 15.5a 15.5a 21 — — 4 BBA5 (4) 2 ≈12.5b 11.8a 13.2a 23 23 1T8J 5 Pin WW domain (5) 2 9.1 9.3a 4.2a 36 34 1PIN (6–39) 6 HP36 (6) 2 10.6 –c –c 36 36 1VII 7 FBP28 (7) 2 9.2 10.6 8.6 37 37 1E0L 8 WW prototype (7) 2 7.7 8.9 7.1 38 37 1E0M 9 psbd41 (8) 2 9.8 –c –c 41 41 2PDD 10 YAP65 (7) 2 7.0 8.4 6.7 40 40 1JMQ (5–44) 11 c-Myb (9) 2 3.1 8.7 1.7 55 54 1IDY 12 1prb (10) 2 13.3 –c –c 47 47 1PRB (7–53) E3-binding domain 13 2 8.0 11.2 6.6 47 47 2WAV of BBL (11) 14 LysM domain (12) 2 3.7 7.0 2.3 66 48 1E0G 15 hTRF1 (9) 2 1.6 5.9 1.2 53 53 1BA5 16 POB (13) 2 8.0 12.3 6.3 51 51 1W4J 17 En-HD (9) >2 8.1 10.5 7.6 54 54 1ENH 18 NTL9 (14) 2 1.9 6.6 0.1 56 56 1DIV (1–56) B1 domain of 19 2 -0.4 6.3 -1.7 62 56 1PGB protein G (14) 20 Src SH3 (14) 2 0.4 4.4 -1.3 61 56 1SRM 21 Fyn SH3 (14) 2 -2.0 4.9 -4.3 78 57 1AVZ (chain C) 22 Spectrin SH3 (14) 2 -3.7 1.1 -4.8 62 57 1SHG Pit1 homeodomain 23 >2 7.4 9.7 4.8 63 58 1AU7 (103–160) (15) 24 Abp1 SH3 (14) 2 -2.1 2.5 -2.7 68 58 1JO8 25 hRAP1 (9) 2 3.9 8.2 2.9 60 59 1FEX 26 BdpA (16) 2 5.8 11.7 4.2 58 58 1BDD (2–59) B1 domain of 27 2 -1.4 4.1 -3.3 72 60 2PTL (18–77) protein L (14) 28 Sso7d (17) 2 0.6 7.0 -3.2 64 63 1BF4 29 Ci2 (14) 2 -3.5 5.8 -10.3 64 64 2CI2 30 434 Cro (18) >2 0.3 3.7 -0.5 71 65 2CRO CspB (B. 31 2 0.2 7.2 -0.4 66 66 1C9O caldolyticus) (19) CspB (T. maritima) 32 2 -2.6 6.3 -4.0 66 66 1G6P (19) CspB (B. subtilis) 33 2 2.7 6.5 2.3 67 67 1CSP (19) 34 EC298 (14) 2 6.4 9.1 4.5 89 69 1JYG 35 CspA (20) 2 1.6 5.3 1.4 69 69 1MJC
1 36 FF domain (21) >2 2.3 7.6 1.4 71 71 1UZC 37 α3D (22) 2 12.4 –c –c 73 73 2A3D 38 Ubiquitin (14) 2 -1.5 7.3 -6.8 76 76 1UBQ 39 GW1 (14) 2 0.1 4.0 -1.7 85 76 1M9S 40 raf RBD (14) 2 -0.2 8.4 -2.8 80 78 1RFA 41 l-repressor (14) 2 5.2 10.4 3.2 80 80 1LMB 42 ADAh2 (14) 2 1.5 6.8 -0.4 81 81 1O6X C-domain of spore 43 2 -8.1 -2.0 -9.3 83 83 1PRS (91–173) coat protein S (23) apocytochrome b 44 5 2 0.1 3.0 -1.4 84 84 1EHB (24) 45 Im7* (14) 2 2.8 7.2 2.3 94 85 1AYI 46 Im9* (14) 2 -1.4 7.3 -1.9 93 85 1IMQ 47 HPr (25) 2 -3.0 2.7 -6.2 85 85 1POH 48 ACBP (Cow) (14) 2 -0.3 7.0 -3.9 86 86 1NTI 49 ACBP (Yeast) (26) >2 -0.4 8.5 6.4 86 86 1ST7 CAfn2 (B. 50 2 -3.8 1.4 -7.0 88 86 1K85 (559–644) circulans) (27) 51 PI3-SH3 (28) 2 -4.8 -1.0 -7.3 90 86 1PNJ 52 HypF-N (29) >2 -0.8 4.4 -6.8 91 88 1GXT 53 Barstar (30) >2 -1.4 3.4 -2.7 89 89 1BTB 54 TNfn3 (31) 2 -4.4 1.1 -7.7 90 89 1TEN 55 TI I27 (32) >2 -6.9 3.6 -7.6 89 89 1TIU N-domain of spore 56 2 -4.6 3.0 -9.4 90 90 1PRS (1–90) coat protein S (23) 1FNF (1326– 57 9FNIII (33) 2 -2.2 -0.9 -2.9 90 90 1415) 1DIV 58 CTL9 (14) 2 -4.4 3.3 -7.9 92 92 (58–149) 59 TWIg18' (34) 2 -5.9 0.4 -8.2 93 93 1WIT 60 hbLBD (35) 2 -4.7 -0.9 -7.8 93 93 1K8M Third PDZ domain 61 >2 -0.2 3.0 -3.4 100 93 1BFE from PSD-95 (36) 62 FNfn10 (37) >2 -0.5 5.5 -8.4 94 94 1TTF PDZ2 domain 63 >2 -1.2 0.8 -2.3 94 94 1GM1 (9–102) from PTP-BL (38) 64 FRB (39) >2 -2.4 6.0 -5.4 97 94 1AUE 65 CD2.D1 (40) >2 -3.5 1.8 -7.6 98 98 1HNG (2–99) 66 L23 (14) 2 -2.4 2.0 -3.9 96 96 1N88 67 U1A (14) 2 -0.4 4.6 -11.7 102 96 1URN 68 suc1(41) >2 -1.0 4.2 -6.1 113 96 1SCE 69 S6 (14) 2 -3.9 6.1 -8.3 101 97 1RIS 70 muscle AcP (14) 2 -7.3d -1.6d -9.0d 99 98 1APS common-type AcP 71 2 -4.4 0.8 -6.5 98 98 2ACY (42) 72 SrcSH2 (14) 2 -1.4 8.7 -3.5 110 103 1SPR 73 Death domain (43) 2 -0.6 6.8 -3.0 107 100 1E41 (93–192) 74 R16 (44) 2 -2.0 4.8 -6.0 116 106 1CUN (7–112) apocytochrome b 75 562 2 -0.1 12.3 -5.0 106 106 256B (45) 1CUN 76 R17 (44) 2 -3.4 3.4 -7.8 116 107 (113–219)
2 77 FKBP (14) 2 -5.2 1.6 -8.1 110 107 1FKB 78 P13 (46) 2 -8.5 0.0 -11.0 109 109 1QTU (1–109) 79 Barnase (47) >2 -4.3 2.6 -9.1 110 109 1RNB 80 R15 (44) 2 4.0 11.0 0.3 116 110 1U5P 81 Myotrophin (48) >2 -0.3 4.7 -3.2 118 118 2MYO 82 Hisactophilin (49) >2 -6.5 4.0 -9.0 118 118 1HCD 83 Villin 14T (50) >2 -1.6 5.0 -4.1 126 126 2VIL 84 ILBP (51) >2 -2.8 1.3 -4.9 127 127 1EAL 85 CheY (52) >2 -1.3 1.0 -4.4 129 128 3CHY 86 IFABP (53) >2 -2.8 3.4 -4.6 131 131 1IFC 87 CRBPII (53) >2 -5.0 1.4 -9.2 134 133 1OPA 88 CRABPI (53) >2 -6.7 -3.2 -9.8 138 136 1CBI 89 CheW (14) 2 -5.0 7.4 -12.1 151 143 1K0S RNase-H (C. 90 >2 -4 1.9 -14 146 146 3H08 tepidum) (54) Apoflavodoxin (D. 91 >2 -4.0 3.6 -4.8 148 147 3F6R desulfuricans) (55) Staphylococcal 92 >2 -5.3 2.3 -8.4 149 149 1JOO nuclease (56) 93 apoMb (57) >2 -1.4 1.1 -3.8 151 151 1A6N Apical domain of 94 >2 -3.4 -1.5 -5.7 155 155 1AON (191–345) GroEL (58) 95 RNase HI (59) >2 -4.6 0.1 -12.0 155 155 2RN2 96 p16 (60) >2 0.4 3.5 0.2 148 148 2A5E (9–156) 97 p19INK4d (61) >2 -1.9 2.9 -2.3 160 156 1BD8 98 DHFR (62) >2 -5.2 -3.2 -6.1 159 159 1RA9 99 CyPA (63) 2 -3.0 6.6 -10.4 164 164 1LOP 100 T4L (59) >2 -5.7 4.1 -14.0 164 164 2LZM Apoflavodoxin 101 (Anabaena sp.) 2 -0.2 2.8 -2.7 169 168 1FTG (64) 102 N-PGK (65) >2 -1.3 2.3 -4.2 175 175 1PHP (1–175) 103 C-PGK (66) >2 -5.1 -3.9 -10.8 221 219 1PHP (176–394) 104 sIGPS (67) >2 -8.9 -4.6 -13.6 222 222 1IGS (27–248) Carbonic anhydrase 105 >2 -10.6 -4.2 -24.0 260 259 1V9E (68) Trp-synthase α 106 subunit (E. coli) >2 -5.7 -2.5 -8.9 268 267 1QOP (chain A) (69) 107 VlsE (14) 2 -4.7 2.0 -8.5 338 271 1L8W(chain A)
Description of each protein includes: Column 1: “#”, the protein number. Column 2: “Protein short name” with a reference to the original experimental paper on folding kinetics. Column 3: “Number of states”: “2” means “two-state folding” and “>2” means “multi-state folding”, experimentally observed at the lowest studied denaturant concentration. mt mt Column 4: “ln k f =lnku ”, natural logarithm of the experimentally found mid-transition folding (= unfolding) rate constant measured in s-1. w w Columns 5, 6: “ln k f ”, “ ln ku ” are natural logarithms of experimentally found (see references in Column 2) folding and unfolding rate constants under “biologically normal” aqueous
3 w conditions. ln k f is either directly measured in the absence of denaturant, or extrapolated to w “normal” aqueous conditions from the region of experimentally measured folding. ln ku is obtained by linear extrapolation to “normal” aqueous conditions from the region of w w experimentally measured unfolding. ln ku ln k f gives stability of the native state, G/RT. Column 7: “L”, the number of residues in the protein chain. Column 8: “LPDB”, the number of folded residues according to PDB. Column 9: “PDB code”, the protein 3D structure code according to the Protein Data Bank (70). If only a part of the chain was used in experiments on protein folding, this part is designated in the Table.
Notes: a w w for proteins denatured by temperature, ln k f and ln ku values refer to 25°C. b for protein # 4 (BBA5), the experimental mid-transition folding rate was not determined directly (4), since the 3D fold of this protein is unstable even at low temperatures; we take mt w w ln k f 12.5 as the average of ln k f = 11.8 and ln ku = 13.2 found at 25°C. c the point is not taken into account: the experiment was carried out at high temperatures and extrapolation of experimental results to 25°C inevitably contains a large error. d for protein # 70 (muscle AcP), the values mf (ln k f ) / M D and mu (ln ku ) / M D
(where MD is denaturant concentration) were erroneously interchanged in the original paper (14) (K. W. Plaxco, F. Chiti, personal communication); the correct values mt mt w w mf = -4.12 kJ/mol/M and mu = 1.27 kJ/mol/M give ln k f (=lnku ), ln k f , ln ku values presented in the Table.
1. Muñoz V, Thompson PA, Hofrichter J, Eaton WA (1997) Folding dynamics and mechanism of beta-hairpin formation. Nature 390:196–199. 2. Qiu L, Pabit SA, Roitberg AE, Hagen SJ Smaller and faster: the 20-residue Trp-cage protein folds in 4 microseconds. J Am Chem Soc 124:12952–12953 (2002). 3. Thompson PA, Eaton WA, Hofrichter J (1997) Laser temperature jump study of the helix-coil kinetics of an alanine peptide interpreted with a “kinetic zipper” model. Biochemistry 36:9200–9210. 4. Snow CD, Nguyen H, Pande VS, Gruebele M (2002) Absolute comparison of simulated and experimental protein-folding dynamics. Nature 420:102–106. 5. Jager M, Nguyen H, Crane JC, Kelly JW, Gruebele M (2001) The folding mechanism of a beta-sheet: the WW domain. J Mol Biol 311:373–393. 6. Wang M, et al. (2003) Dynamic NMR line-shape analysis demonstrates that the villin headpiece subdomain folds on the microsecond time scale. J Am Chem Soc 125:6032–6033. 7. Ferguson N, Johnson CM, Macias M, Oschkinat H, Fersht A (2001) Ultrafast folding of WW domains without structured aromatic clusters in the denatured state. Proc Natl Acad Sci USA 98:13002–13007. 8. Spector S, Raleigh DP (1999) Submillisecond folding of the peripheral subunit- binding domain. J Mol Biol 293:763–768. 9. Gianni S, et al. (2003) Unifying features in protein-folding mechanisms. Proc Natl Acad Sci USA 100:13286–13291. 10.Wang T, Zhu Y, Gai F (2004) Folding of a three-helix bundle at the folding speed limit. J Phys Chem B 108:3694–3697. 11.Neuweiler H, et al. (2009) The folding mechanism of BBL: plasticity of transition- state structure observed within an ultrafast folding protein family. J Mol Biol 390:1060–1073.
4 12.Nickson AA, Stoll KE, Clarke J (2008) Folding of a LysM domain: entropy-enthalpy compensation in the transition state of an ideal two-state folder. J Mol Biol 380:557– 569. 13.Sharpe TD, Ferguson N, Johnson CM, Fersht AR (2008) Conservation of transition state structure in fast folding peripheral subunit-binding domains. J Mol Biol 383:224–237. 14.Maxwell KL, et al. (2005) Protein folding: defining a “standard” set of experimental conditions and a preliminary kinetic data set of two-state proteins. Protein Sci 14:602–616. 15.Banachewicz W, Johnson CM, Fersht AR (2011) Folding of the Pit1 homeodomain near the speed limit. Proc Natl Acad Sci USA 108:569–573. 16.Myers JK, Oas TG (2001) Preorganized secondary structure as an important determinant of fast protein folding. Nat Struct Biol 8:552–558. 17.Guerois R, Serrano L (2000) The SH3-fold family: experimental evidence and prediction of variations in the folding pathways. J Mol Biol 304:967–982. 18.Laurents DV, et al. (2000) Folding kinetics of phage 434 Cro protein. Biochemistry 39:13963–13973. 19.Perl D, et al. (1998) Conservation of rapid two-state folding in mesophilic, thermophilic and hyperthermophilic cold shock proteins. Nat Struct Biol 5:229–235. 20.Reid KL, Rodriguez HM, Hillier BJ, Gregoret LM (1998) Stability and folding properties of a model beta-sheet protein, Escherichia coli CspA. Protein Sci 7:470– 479. 21.Jemth P, et al. (2005) The structure of the major transition state for folding of an FF domain from experiment and simulation. J Mol Biol 350:363–378. 22.Zhu Y, et al. (2003) Ultrafast folding of alpha3D: a de novo designed three-helix bundle protein. Proc Natl Acad Sci USA 100:15486–15491. 23.Wenk M, Jaenicke R, Mayr E (1998) Kinetic stabilisation of a modular protein by domain interactions. FEBS Lett 438:127–130. 24.Manyusa S, Whitford D (1999) Defining folding and unfolding reactions of apocytochrome b5 using equilibrium and kinetic fluorescence measurements. Biochemistry 38:9533–9540. 25.van Nuland NA, et al. (1998) Slow cooperative folding of a small globular protein HPr. Biochemistry 37:622–637. 26.Teilum K, et al. (2005) Different secondary structure elements as scaffolds for protein folding transition states of two homologous four-helix bundles. Proteins 59:80–90. 27.Lappalainen I, Hurley MG, Clarke J (2008) Plasticity within the obligatory folding nucleus of an immunoglobulin-like domain. J Mol Biol 375:547–559. 28.Guijarro JI, Morton CJ, Plaxco KW, Campbell ID, Dobson CM (1998) Folding kinetics of the SH3 domain of PI3 kinase by real-time NMR combined with optical spectroscopy. J Mol Biol 276:657–667. 29.Calloni G, et al. (2003) Comparison of the folding processes of distantly related proteins. Importance of hydrophobic content in folding. J Mol Biol 330:577–591. 30.Schreiber G, Fersht AR (1993) The refolding of cis- and trans-peptidylprolyl isomers of barstar. Biochemistry 32:11195–11203. 31.Clarke J, Hamill SJ, Johnson CM (1997) Folding and stability of a fibronectin type III domain of human tenascin. J Mol Biol 270:771–778. 32.Fowler SB, Clarke J (2001) Mapping the folding pathway of an immunoglobulin domain: structural detail from phi value analysis and movement of the transition state. Structure 9:355–366.
5 33.Plaxco KW, Spitzfaden C, Campbell ID, Dobson CM (1997) A comparison of the folding kinetics and thermodynamics of two homologous fibronectin type III modules. J Mol Biol 270:763–770. 34.Clarke J, Cota E, Fowler SB, Hamill SJ (1999) Folding studies of immunoglobulin- like beta-sandwich proteins suggest that they share a common folding pathway. Structure 7:1145–1153. 35.Naik MT, Chang YC, Huang TH (2002) Folding kinetics of the lipoic acid-bearing domain of human mitochondrial branched chain alpha-ketoacid dehydrogenase complex. FEBS Lett 530:133–138. 36.Calosci N, et al. (2008) Comparison of successive transition states for folding reveals alternative early folding pathways of two homologous proteins. Proc Natl Acad Sci USA 105:19241–19246. 37.Cota E, Clarke J (2000) Folding of beta-sandwich proteins: three-state transition of a fibronectin type III module. Protein Sci 9:112–120. 38.Gianni S, et al. (2005) Kinetic folding mechanism of PDZ2 from PTP-BL. Prot Eng Des Sel 18:389–395. 39.Marianayagam NJ, Khan F, Male L, Jackson SE (2002) Fast folding of a four-helical bundle protein. J Am Chem Soc 124:9744–9750. 40.Parker MJ, Dempsey CE, Lorch M, Clarke AR (1997) Acquisition of native beta- strand topology during the rapid collapse phase of protein folding. Biochemistry 36:13396–13405. 41.Schymkowitz JW, Rousseau F, Irvine LR, Itzhaki LS (2000) The folding pathway of the cell-cycle regulatory protein p13suc1: clues for the mechanism of domain swapping. Structure 8:89–100. 42.Taddei N, et al. (1999) Thermodynamics and kinetics of folding of common-type acylphosphatase: comparison to the highly homologous muscle isoenzyme. Biochemistry 38:2135–2142. 43.Steward A, McDowell GS, Clarke J (2009) Topology is the principal determinant in the folding of a complex all-alpha greek key death domain from human FADD. J Mol Biol 389:425–437. 44.Scott KA, Batey S, Hooton KA, Clarke J (2004) The folding of spectrin domains I: wild-type domains have the same stability but very different kinetic properties. J Mol Biol 344:195–205. 45.Wittung-Stafshede P, Lee JC, Winkler JR, Gray HB (1999) Cytochrome b562 folding triggered by electron transfer: approaching the speed limit for formation of a four- helix-bundle protein. Proc Natl Acad Sci USA 96:6587–6590. 46.Roumestand C, Boyer M, Guignard L, Barthe P, Royer CA (2001) Characterization of the folding and unfolding reactions of a small beta-barrel protein of novel topology, the MTCP1 oncogene product P13. J Mol Biol 312:247–259. 47.Matouschek A, Kellis JT Jr, Serrano L, Bycroft M, Fersht, AR (1990) Transient folding intermediates characterized by protein engineering. Nature 346:440–445. 48.Lowe AR, Itzhaki LS (2007) Rational redesign of the folding pathway of a modular protein. Proc Natl Acad Sci USA 104:2679–2684. 49.Liu C, Gaspar JA, Wong HJ, Meiering EM (2002) Conserved and nonconserved features of the folding pathway of hisactophilin, a beta-trefoil protein. Protein Sci 11:669–679. 50.Choe SE, Matsudaira PT, Osterhout J, Wagner G, Shakhnovich EI (1998) Folding kinetics of villin 14T, a protein domain with a central beta-sheet and two hydrophobic cores. Biochemistry 37:14508–14518. 51.Dalessio PM, Ropson IJ (2000) Beta-sheet proteins with nearly identical structures have different folding intermediates. Biochemistry 39:860–871.
6 52.Muñoz V, Lopez EM, Jager M, Serrano L (1994) Kinetic characterization of the chemotactic protein from Escherichia coli, CheY. Kinetic analysis of the inverse hydrophobic effect. Biochemistry 33:5858–5866. 53.Burns LL, Dalessio PM, Ropson IJ (1998) Folding mechanism of three structurally similar beta-sheet proteins. Proteins 33:107–118. 54.Ratcliff K, Corn J, Marqusee S (2009) Structure, stability, and folding of ribonuclease H1 from the moderately thermophilic Chlorobium tepidum: comparison with thermophilic and mesophilic homologues. Biochemistry 48:5890–5898. 55.Stagg L, Samiotakis A, Homouz D, Cheung MS, Wittung-Stafshede P (2010) Residue-specific analysis of frustration in the folding landscape of repeat beta/alpha protein apoflavodoxin. J Mol Biol 396:75–89. 56.Maki K, Cheng H, Dolgikh DA, Shastry MC, Roder H (2004) Early events during folding of wild-type staphylococcal nuclease and a single-tryptophan variant studied by ultrarapid mixing. J Mol Biol 338:383–400. 57.Cavagnero S, Dyson HJ, Wright PE (1999) Effect of H helix destabilizing mutations on the kinetic and equilibrium folding of apomyoglobin. J Mol Biol 285:269–282. 58.Golbik R, Zahn R, Harding SE, Fersht AR (1998) Thermodynamic stability and folding of GroEL minichaperones. J Mol Biol 276:505–515. 59.Parker MJ, Marqusee S (1999) The cooperativity of burst phase reactions explored. J Mol Biol 293:1195–1210. 60.Tang KS, Guralnick BJ, Wang WK, Fersht AR, Itzhaki, LS (1999) Stability and folding of the tumour suppressor protein p16. J Mol Biol 285:1869–1886. 61.Löw C, et al. (2007) Folding mechanism of an ankyrin repeat protein: scaffold and active site formation of human CDK inhibitor p19INK4d. J Mol Biol 373:219–231. 62.Jennings PA, Finn BE, Jones BE, Matthews CR (1993) A reexamination of the folding mechanism of dihydrofolate reductase from Escherichia coli: verification and refinement of a four-channel model. Biochemistry 32:3783–3789. 63.Ikura T, Hayano T, Takahashi N, Kuwajima K (2000) Fast folding of Escherichia coli cyclophilin A: a hypothesis of a unique hydrophobic core with a phenylalanine cluster. J Mol Biol 297:791–802. 64.Bueno M, Ayuso-Tejedor S, Sancho J (2006) Do proteins with similar folds have similar transition state structures? A diffuse transition state of the 169 residue apoflavodoxin. J Mol Biol 359:813–824. 65.Parker MJ, Spencer J, Clarke AR (1995) An integrated kinetic analysis of intermediates and transition states in protein folding reactions. J Mol Biol 253:771– 786. 66.Parker MJ, et al. (1996) Domain behavior during the folding of a thermostable phosphoglycerate kinase. Biochemistry 35:15740–15752. 67.Forsyth WR, Matthews CR (2002) Folding mechanism of indole-3-glycerol phosphate synthase from Sulfolobus solfataricus: a test of the conservation of folding mechanisms hypothesis in (beta-alpha)8 barrels. J Mol Biol 320:1119–1133. 68.Melnik BS, Marchenkov VV, Evdokimov SR, Samatova EN, Kotova NV (2008) Multy- state protein: determination of carbonic anhydrase free-energy landscape. Biochem Biophys Res Commun 369:701–706. 69.Ogasahara K, Yutani K (1994) Unfolding-refolding kinetics of the tryptophan synthase alpha subunit by CD and fluorescence measurements. J Mol Biol 236:1227–1240. 70.Berman HM, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242.
7